Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
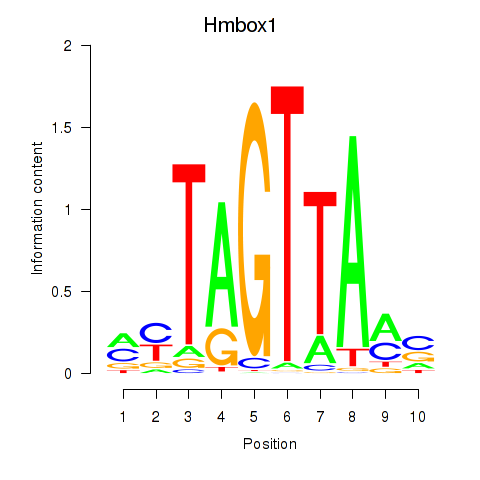
Results for Hmbox1
Z-value: 0.91
Transcription factors associated with Hmbox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmbox1
|
ENSMUSG00000021972.8 | homeobox containing 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmbox1 | mm10_v2_chr14_-_64949632_64949674 | 0.25 | 1.3e-01 | Click! |
Activity profile of Hmbox1 motif
Sorted Z-values of Hmbox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39287074 | 8.65 |
ENSMUST00000003137.8
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr14_+_37068042 | 4.95 |
ENSMUST00000057176.3
|
Lrit2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr11_-_78422217 | 4.33 |
ENSMUST00000001122.5
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr10_-_115362191 | 2.61 |
ENSMUST00000092170.5
|
Tmem19
|
transmembrane protein 19 |
| chr8_+_45507768 | 2.60 |
ENSMUST00000067065.7
ENSMUST00000098788.3 ENSMUST00000067107.7 ENSMUST00000171337.2 ENSMUST00000138049.1 ENSMUST00000141039.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr11_-_95041335 | 2.53 |
ENSMUST00000038431.7
|
Pdk2
|
pyruvate dehydrogenase kinase, isoenzyme 2 |
| chr18_-_38866702 | 2.49 |
ENSMUST00000115582.1
|
Fgf1
|
fibroblast growth factor 1 |
| chr8_+_45658666 | 2.44 |
ENSMUST00000134675.1
ENSMUST00000139869.1 ENSMUST00000126067.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr13_+_4434306 | 2.24 |
ENSMUST00000021630.8
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr15_+_55112420 | 2.23 |
ENSMUST00000100660.4
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr14_+_66635251 | 2.22 |
ENSMUST00000159365.1
ENSMUST00000054661.1 ENSMUST00000159068.1 |
Adra1a
|
adrenergic receptor, alpha 1a |
| chr18_-_3281036 | 2.17 |
ENSMUST00000049942.6
ENSMUST00000139537.1 ENSMUST00000124747.1 |
Crem
|
cAMP responsive element modulator |
| chr3_-_116711820 | 2.12 |
ENSMUST00000153108.1
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr8_+_45999297 | 2.07 |
ENSMUST00000110380.1
ENSMUST00000066451.3 |
Lrp2bp
|
Lrp2 binding protein |
| chr15_+_25773985 | 2.04 |
ENSMUST00000125667.1
|
Myo10
|
myosin X |
| chr4_+_100478806 | 1.99 |
ENSMUST00000133493.2
ENSMUST00000092730.3 ENSMUST00000106979.3 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr5_+_87000838 | 1.90 |
ENSMUST00000031186.7
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr15_+_55112317 | 1.87 |
ENSMUST00000096433.3
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr8_+_45658731 | 1.80 |
ENSMUST00000143820.1
ENSMUST00000132139.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr19_+_5038826 | 1.62 |
ENSMUST00000053705.6
|
B3gnt1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chrM_+_5319 | 1.57 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr11_+_101468164 | 1.53 |
ENSMUST00000001347.6
|
Rnd2
|
Rho family GTPase 2 |
| chr7_-_45061706 | 1.52 |
ENSMUST00000107832.1
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr1_+_21240597 | 1.50 |
ENSMUST00000121676.1
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr11_-_49114874 | 1.47 |
ENSMUST00000109201.1
|
Olfr1396
|
olfactory receptor 1396 |
| chr1_+_21240581 | 1.46 |
ENSMUST00000027067.8
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr2_-_12419387 | 1.46 |
ENSMUST00000124515.1
|
Fam188a
|
family with sequence similarity 188, member A |
| chr4_+_99030946 | 1.42 |
ENSMUST00000030280.6
|
Angptl3
|
angiopoietin-like 3 |
| chr4_+_116558056 | 1.39 |
ENSMUST00000106475.1
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr11_+_93996082 | 1.38 |
ENSMUST00000041956.7
|
Spag9
|
sperm associated antigen 9 |
| chr6_+_29853746 | 1.27 |
ENSMUST00000064872.6
ENSMUST00000152581.1 ENSMUST00000176265.1 ENSMUST00000154079.1 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr3_-_79842662 | 1.25 |
ENSMUST00000029568.1
|
Tmem144
|
transmembrane protein 144 |
| chr13_-_23574196 | 1.25 |
ENSMUST00000105106.1
|
Hist1h2bf
|
histone cluster 1, H2bf |
| chr1_-_131279544 | 1.18 |
ENSMUST00000062108.3
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr1_+_59256906 | 1.16 |
ENSMUST00000160662.1
ENSMUST00000114248.2 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr2_+_3770673 | 1.15 |
ENSMUST00000177037.1
|
Fam107b
|
family with sequence similarity 107, member B |
| chr7_-_45061651 | 1.15 |
ENSMUST00000007981.3
ENSMUST00000107831.1 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr12_+_64965742 | 1.14 |
ENSMUST00000066296.7
|
Fam179b
|
family with sequence similarity 179, member B |
| chr11_-_88851462 | 1.12 |
ENSMUST00000107903.1
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr17_+_40934676 | 1.11 |
ENSMUST00000169611.2
|
Mut
|
methylmalonyl-Coenzyme A mutase |
| chr15_+_99393610 | 1.10 |
ENSMUST00000159531.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr16_+_35983424 | 1.08 |
ENSMUST00000173555.1
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr2_+_34874396 | 1.07 |
ENSMUST00000113068.2
ENSMUST00000047447.8 |
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr3_+_108653979 | 1.05 |
ENSMUST00000106613.1
|
Clcc1
|
chloride channel CLIC-like 1 |
| chr16_+_10545390 | 1.05 |
ENSMUST00000115827.1
ENSMUST00000038145.6 ENSMUST00000150894.1 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr3_+_108653931 | 1.01 |
ENSMUST00000029483.8
ENSMUST00000124384.1 |
Clcc1
|
chloride channel CLIC-like 1 |
| chr15_+_99393574 | 1.01 |
ENSMUST00000162624.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr7_-_109723786 | 1.01 |
ENSMUST00000106735.2
ENSMUST00000033334.4 |
BC051019
|
cDNA sequence BC051019 |
| chr1_+_156040884 | 0.99 |
ENSMUST00000060404.4
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr19_-_28967794 | 0.99 |
ENSMUST00000162110.1
|
4430402I18Rik
|
RIKEN cDNA 4430402I18 gene |
| chr12_-_81532840 | 0.99 |
ENSMUST00000169158.1
ENSMUST00000164431.1 ENSMUST00000163402.1 ENSMUST00000166664.1 ENSMUST00000164386.1 |
Synj2bp
Gm20498
|
synaptojanin 2 binding protein predicted gene 20498 |
| chr4_+_20042046 | 0.98 |
ENSMUST00000098242.3
|
Ggh
|
gamma-glutamyl hydrolase |
| chr2_+_34874486 | 0.94 |
ENSMUST00000028228.3
|
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr10_+_22158566 | 0.93 |
ENSMUST00000181645.1
ENSMUST00000105522.2 |
Raet1e
H60b
|
retinoic acid early transcript 1E histocompatibility 60b |
| chr3_+_108653907 | 0.93 |
ENSMUST00000106609.1
|
Clcc1
|
chloride channel CLIC-like 1 |
| chr11_+_51763682 | 0.92 |
ENSMUST00000020653.5
|
Sar1b
|
SAR1 gene homolog B (S. cerevisiae) |
| chr12_-_113361232 | 0.90 |
ENSMUST00000103423.1
|
Ighg3
|
Immunoglobulin heavy constant gamma 3 |
| chr1_-_16520020 | 0.87 |
ENSMUST00000144138.2
ENSMUST00000145092.1 |
Stau2
|
staufen (RNA binding protein) homolog 2 (Drosophila) |
| chr1_+_88200601 | 0.86 |
ENSMUST00000049289.8
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr4_-_133339283 | 0.85 |
ENSMUST00000043305.7
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr9_+_64235201 | 0.85 |
ENSMUST00000039011.3
|
Uchl4
|
ubiquitin carboxyl-terminal esterase L4 |
| chr17_+_13007839 | 0.84 |
ENSMUST00000007012.4
|
Sod2
|
superoxide dismutase 2, mitochondrial |
| chr2_-_157571270 | 0.84 |
ENSMUST00000173378.1
|
Blcap
|
bladder cancer associated protein homolog (human) |
| chr13_-_3893556 | 0.83 |
ENSMUST00000099946.4
|
Net1
|
neuroepithelial cell transforming gene 1 |
| chr4_-_133339238 | 0.82 |
ENSMUST00000105906.1
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr9_+_75410145 | 0.80 |
ENSMUST00000180533.1
ENSMUST00000180574.1 |
4933433G15Rik
|
RIKEN cDNA 4933433G15 gene |
| chr16_+_10545339 | 0.79 |
ENSMUST00000066345.7
ENSMUST00000115824.3 ENSMUST00000155633.1 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr9_-_71168657 | 0.77 |
ENSMUST00000113570.1
|
Aqp9
|
aquaporin 9 |
| chr6_+_113638467 | 0.77 |
ENSMUST00000059286.7
ENSMUST00000089023.4 ENSMUST00000089022.4 |
Irak2
|
interleukin-1 receptor-associated kinase 2 |
| chr14_+_55971428 | 0.75 |
ENSMUST00000089555.2
|
Cma2
|
chymase 2, mast cell |
| chr4_+_128993224 | 0.75 |
ENSMUST00000030583.6
ENSMUST00000102604.4 |
Ak2
|
adenylate kinase 2 |
| chr3_+_5214763 | 0.74 |
ENSMUST00000177924.1
|
Gm10748
|
predicted gene 10748 |
| chr5_-_136565432 | 0.74 |
ENSMUST00000176172.1
|
Cux1
|
cut-like homeobox 1 |
| chr4_-_58912678 | 0.74 |
ENSMUST00000144512.1
ENSMUST00000102889.3 ENSMUST00000055822.8 |
AI314180
|
expressed sequence AI314180 |
| chr1_-_16520067 | 0.73 |
ENSMUST00000131257.2
ENSMUST00000153966.2 |
Stau2
|
staufen (RNA binding protein) homolog 2 (Drosophila) |
| chr12_+_17266545 | 0.73 |
ENSMUST00000057288.5
|
Pdia6
|
protein disulfide isomerase associated 6 |
| chr3_-_123236134 | 0.72 |
ENSMUST00000106427.1
ENSMUST00000106426.1 ENSMUST00000051443.5 |
Synpo2
|
synaptopodin 2 |
| chr18_+_32938955 | 0.72 |
ENSMUST00000042868.4
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr12_-_64965496 | 0.70 |
ENSMUST00000021331.7
|
Klhl28
|
kelch-like 28 |
| chr8_-_9977650 | 0.69 |
ENSMUST00000170033.1
|
Lig4
|
ligase IV, DNA, ATP-dependent |
| chr19_-_4928241 | 0.69 |
ENSMUST00000025851.3
|
Dpp3
|
dipeptidylpeptidase 3 |
| chr7_-_29726991 | 0.67 |
ENSMUST00000061193.3
|
Catsperg2
|
catsper channel auxiliary subunit gamma 2 |
| chr13_+_74406387 | 0.64 |
ENSMUST00000090860.6
|
Gm10116
|
predicted pseudogene 10116 |
| chr1_-_16520106 | 0.60 |
ENSMUST00000162435.1
|
Stau2
|
staufen (RNA binding protein) homolog 2 (Drosophila) |
| chr12_-_69893162 | 0.59 |
ENSMUST00000049239.7
ENSMUST00000110570.1 |
Map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr2_-_34826187 | 0.58 |
ENSMUST00000113075.1
ENSMUST00000113080.2 ENSMUST00000091020.3 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr13_-_74807913 | 0.58 |
ENSMUST00000065629.4
|
Cast
|
calpastatin |
| chr1_+_15805639 | 0.57 |
ENSMUST00000027057.6
|
Terf1
|
telomeric repeat binding factor 1 |
| chr6_+_96115249 | 0.56 |
ENSMUST00000075080.5
|
Fam19a1
|
family with sequence similarity 19, member A1 |
| chr15_-_58034289 | 0.56 |
ENSMUST00000100655.3
|
9130401M01Rik
|
RIKEN cDNA 9130401M01 gene |
| chr3_+_145118564 | 0.55 |
ENSMUST00000098538.2
ENSMUST00000106192.2 ENSMUST00000029920.8 ENSMUST00000098539.2 |
Odf2l
|
outer dense fiber of sperm tails 2-like |
| chr2_+_175010241 | 0.54 |
ENSMUST00000109069.1
ENSMUST00000109070.2 |
Gm14444
|
predicted gene 14444 |
| chr14_+_53795455 | 0.54 |
ENSMUST00000103671.2
|
B230359F08Rik
|
RIKEN cDNA B230359F08 gene |
| chr5_+_104170676 | 0.53 |
ENSMUST00000112771.1
|
Dspp
|
dentin sialophosphoprotein |
| chr9_-_104102550 | 0.53 |
ENSMUST00000050139.4
|
Ackr4
|
atypical chemokine receptor 4 |
| chr6_-_97179100 | 0.52 |
ENSMUST00000095664.3
|
Tmf1
|
TATA element modulatory factor 1 |
| chr18_+_65698253 | 0.51 |
ENSMUST00000115097.1
ENSMUST00000117694.1 |
Oacyl
|
O-acyltransferase like |
| chr8_+_33516730 | 0.50 |
ENSMUST00000149399.1
|
Tex15
|
testis expressed gene 15 |
| chr17_+_24023859 | 0.47 |
ENSMUST00000178572.1
|
Gm5225
|
predicted gene 5225 |
| chr13_-_34077992 | 0.46 |
ENSMUST00000056427.8
|
Tubb2a
|
tubulin, beta 2A class IIA |
| chr4_-_150914401 | 0.46 |
ENSMUST00000105675.1
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr17_-_12851893 | 0.45 |
ENSMUST00000162389.1
ENSMUST00000162119.1 ENSMUST00000159223.1 |
Mas1
|
MAS1 oncogene |
| chrY_+_80135210 | 0.44 |
ENSMUST00000179811.1
|
Gm21760
|
predicted gene, 21760 |
| chr5_+_7304128 | 0.44 |
ENSMUST00000160634.1
ENSMUST00000159546.1 |
4921511H03Rik
|
RIKEN cDNA 4921511H03 gene |
| chr3_+_5218589 | 0.43 |
ENSMUST00000177488.1
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr18_-_6241486 | 0.42 |
ENSMUST00000025083.7
|
Kif5b
|
kinesin family member 5B |
| chrX_-_140062712 | 0.41 |
ENSMUST00000133780.1
|
Nup62cl
|
nucleoporin 62 C-terminal like |
| chr1_-_31222604 | 0.41 |
ENSMUST00000127775.1
|
4931428L18Rik
|
RIKEN cDNA 4931428L18 gene |
| chr12_-_112802646 | 0.40 |
ENSMUST00000124526.1
|
Ahnak2
|
AHNAK nucleoprotein 2 |
| chr8_+_110079758 | 0.40 |
ENSMUST00000058804.8
|
Zfp612
|
zinc finger protein 612 |
| chr17_+_56079652 | 0.40 |
ENSMUST00000002911.8
|
Hdgfrp2
|
hepatoma-derived growth factor, related protein 2 |
| chr6_+_33249085 | 0.39 |
ENSMUST00000052266.8
ENSMUST00000090381.4 ENSMUST00000115080.1 |
Exoc4
|
exocyst complex component 4 |
| chr13_+_55209776 | 0.39 |
ENSMUST00000099490.2
|
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr1_-_97761538 | 0.39 |
ENSMUST00000171129.1
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr12_+_69893105 | 0.39 |
ENSMUST00000021466.8
|
Atl1
|
atlastin GTPase 1 |
| chr10_-_81600857 | 0.39 |
ENSMUST00000151858.1
ENSMUST00000142948.1 ENSMUST00000072020.2 |
Tle6
|
transducin-like enhancer of split 6, homolog of Drosophila E(spl) |
| chr1_-_191907527 | 0.39 |
ENSMUST00000069573.5
|
1700034H15Rik
|
RIKEN cDNA 1700034H15 gene |
| chr3_-_57294880 | 0.39 |
ENSMUST00000171384.1
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr16_-_48771956 | 0.39 |
ENSMUST00000170861.1
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr6_-_42597372 | 0.38 |
ENSMUST00000069023.3
|
Fam115e
|
family with sequence similarity 115, member E |
| chr10_-_40257648 | 0.38 |
ENSMUST00000019982.7
|
Gtf3c6
|
general transcription factor IIIC, polypeptide 6, alpha |
| chr4_+_152338887 | 0.38 |
ENSMUST00000005175.4
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr5_+_90561102 | 0.37 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr7_+_83891983 | 0.36 |
ENSMUST00000094215.3
ENSMUST00000130103.1 |
Mesdc2
|
mesoderm development candidate 2 |
| chr9_+_104566677 | 0.36 |
ENSMUST00000157006.1
|
Cpne4
|
copine IV |
| chr12_-_31634592 | 0.36 |
ENSMUST00000020979.7
ENSMUST00000177962.1 |
Bcap29
|
B cell receptor associated protein 29 |
| chr18_-_61211572 | 0.35 |
ENSMUST00000146409.1
|
Slc26a2
|
solute carrier family 26 (sulfate transporter), member 2 |
| chr8_-_88362154 | 0.35 |
ENSMUST00000034085.7
|
Brd7
|
bromodomain containing 7 |
| chr3_-_108889990 | 0.34 |
ENSMUST00000053065.4
ENSMUST00000102620.3 |
Fndc7
|
fibronectin type III domain containing 7 |
| chr2_+_155276297 | 0.34 |
ENSMUST00000029128.3
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr6_+_18866309 | 0.34 |
ENSMUST00000115396.2
ENSMUST00000031489.6 |
Ankrd7
|
ankyrin repeat domain 7 |
| chr18_-_6241470 | 0.34 |
ENSMUST00000163210.1
|
Kif5b
|
kinesin family member 5B |
| chr3_+_95588990 | 0.34 |
ENSMUST00000177399.1
|
Golph3l
|
golgi phosphoprotein 3-like |
| chr3_+_118433797 | 0.33 |
ENSMUST00000180593.1
ENSMUST00000181926.1 ENSMUST00000181060.1 ENSMUST00000181310.1 |
Gm26871
|
predicted gene, 26871 |
| chr11_-_73177002 | 0.33 |
ENSMUST00000108480.1
ENSMUST00000054952.3 |
Emc6
|
ER membrane protein complex subunit 6 |
| chr10_+_112271123 | 0.32 |
ENSMUST00000092175.2
|
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr4_-_119174178 | 0.32 |
ENSMUST00000106355.3
|
Zfp691
|
zinc finger protein 691 |
| chr10_-_11080956 | 0.32 |
ENSMUST00000105560.1
|
Grm1
|
glutamate receptor, metabotropic 1 |
| chr2_-_130284422 | 0.32 |
ENSMUST00000028892.4
|
Idh3b
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr11_+_23008337 | 0.31 |
ENSMUST00000172602.1
|
Fam161a
|
family with sequence similarity 161, member A |
| chr1_+_156040909 | 0.31 |
ENSMUST00000065648.8
ENSMUST00000097526.2 |
Tor1aip2
|
torsin A interacting protein 2 |
| chr7_-_131146306 | 0.30 |
ENSMUST00000070980.3
|
4933402N03Rik
|
RIKEN cDNA 4933402N03 gene |
| chr3_-_95217741 | 0.29 |
ENSMUST00000107204.1
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr1_-_107161114 | 0.29 |
ENSMUST00000086694.4
|
Serpinb3b
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3B |
| chr2_-_169405435 | 0.28 |
ENSMUST00000131509.1
|
4930529I22Rik
|
RIKEN cDNA 4930529I22 gene |
| chr6_-_101377342 | 0.28 |
ENSMUST00000151175.1
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr17_+_69439326 | 0.28 |
ENSMUST00000169935.1
|
A330050F15Rik
|
RIKEN cDNA A330050F15 gene |
| chr14_-_79223876 | 0.27 |
ENSMUST00000040802.4
|
Zfp957
|
zinc finger protein 957 |
| chr3_+_95588960 | 0.26 |
ENSMUST00000176674.1
ENSMUST00000177389.1 ENSMUST00000176755.1 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr6_-_126849109 | 0.26 |
ENSMUST00000088194.4
|
Ndufa9
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9 |
| chr9_-_32928928 | 0.25 |
ENSMUST00000185169.1
|
RP24-308I2.1
|
RP24-308I2.1 |
| chr10_+_39133981 | 0.25 |
ENSMUST00000019991.7
|
Tube1
|
epsilon-tubulin 1 |
| chrX_-_155623118 | 0.23 |
ENSMUST00000170236.1
|
Ptchd1
|
patched domain containing 1 |
| chr2_-_34826071 | 0.23 |
ENSMUST00000113077.1
ENSMUST00000028220.3 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr8_-_54724317 | 0.22 |
ENSMUST00000129132.2
ENSMUST00000150488.1 ENSMUST00000127511.2 |
Wdr17
|
WD repeat domain 17 |
| chr7_-_38528193 | 0.22 |
ENSMUST00000079759.5
|
Gm5591
|
predicted gene 5591 |
| chr4_+_117252010 | 0.22 |
ENSMUST00000125943.1
ENSMUST00000106434.1 |
Tmem53
|
transmembrane protein 53 |
| chr11_+_83942062 | 0.22 |
ENSMUST00000049257.7
|
Ddx52
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 |
| chrX_-_7375830 | 0.21 |
ENSMUST00000115744.1
|
Usp27x
|
ubiquitin specific peptidase 27, X chromosome |
| chrX_+_140062879 | 0.21 |
ENSMUST00000044806.2
|
E230019M04Rik
|
RIKEN cDNA E230019M04 gene |
| chr14_-_70323783 | 0.20 |
ENSMUST00000151011.1
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr11_+_31872100 | 0.20 |
ENSMUST00000020543.6
ENSMUST00000109412.2 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr2_+_35224516 | 0.20 |
ENSMUST00000124489.1
|
Gm13605
|
predicted gene 13605 |
| chr13_-_101692624 | 0.20 |
ENSMUST00000035532.6
|
Pik3r1
|
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha) |
| chr11_+_77686155 | 0.19 |
ENSMUST00000100802.4
ENSMUST00000181023.1 |
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr9_+_36693217 | 0.19 |
ENSMUST00000034620.4
|
Acrv1
|
acrosomal vesicle protein 1 |
| chr13_-_73678005 | 0.18 |
ENSMUST00000022105.7
ENSMUST00000109680.2 ENSMUST00000109679.2 |
Slc6a18
|
solute carrier family 6 (neurotransmitter transporter), member 18 |
| chr12_+_52097737 | 0.17 |
ENSMUST00000040090.9
|
Nubpl
|
nucleotide binding protein-like |
| chr19_-_3282958 | 0.17 |
ENSMUST00000119292.1
ENSMUST00000025751.3 |
Ighmbp2
|
immunoglobulin mu binding protein 2 |
| chr2_+_119208975 | 0.16 |
ENSMUST00000102519.4
|
Zfyve19
|
zinc finger, FYVE domain containing 19 |
| chr3_-_108889706 | 0.16 |
ENSMUST00000180063.1
|
Fndc7
|
fibronectin type III domain containing 7 |
| chr19_-_6235804 | 0.16 |
ENSMUST00000025695.9
|
Ppp2r5b
|
protein phosphatase 2, regulatory subunit B (B56), beta isoform |
| chr1_-_31222644 | 0.15 |
ENSMUST00000135245.1
|
4931428L18Rik
|
RIKEN cDNA 4931428L18 gene |
| chr7_-_140881811 | 0.15 |
ENSMUST00000106048.3
ENSMUST00000147331.2 ENSMUST00000137710.1 |
Sirt3
|
sirtuin 3 |
| chr3_+_93319740 | 0.15 |
ENSMUST00000090856.3
ENSMUST00000093774.3 |
Hrnr
|
hornerin |
| chr11_+_23008420 | 0.14 |
ENSMUST00000109557.2
|
Fam161a
|
family with sequence similarity 161, member A |
| chr3_-_95217877 | 0.13 |
ENSMUST00000136139.1
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chrX_-_140062293 | 0.13 |
ENSMUST00000154385.1
|
Nup62cl
|
nucleoporin 62 C-terminal like |
| chr4_+_149518216 | 0.13 |
ENSMUST00000030839.6
|
Ctnnbip1
|
catenin beta interacting protein 1 |
| chr3_+_95588928 | 0.12 |
ENSMUST00000177390.1
ENSMUST00000098861.4 ENSMUST00000060323.5 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr8_+_40354303 | 0.12 |
ENSMUST00000136835.1
|
Micu3
|
mitochondrial calcium uptake family, member 3 |
| chr11_-_64436653 | 0.12 |
ENSMUST00000177999.1
|
F930015N05Rik
|
RIKEN cDNA F930015N05 gene |
| chr2_+_117249725 | 0.12 |
ENSMUST00000028825.4
|
Fam98b
|
family with sequence similarity 98, member B |
| chr8_+_72099762 | 0.12 |
ENSMUST00000074540.4
|
Olfr373
|
olfactory receptor 373 |
| chr19_+_23675839 | 0.11 |
ENSMUST00000056396.5
|
Gm6563
|
predicted pseudogene 6563 |
| chr9_-_83254460 | 0.11 |
ENSMUST00000184080.1
ENSMUST00000184100.1 |
RP23-341H6.1
|
RP23-341H6.1 |
| chr7_-_29213957 | 0.11 |
ENSMUST00000169143.1
ENSMUST00000047846.6 |
Catsperg1
|
catsper channel auxiliary subunit gamma 1 |
| chr3_-_123690806 | 0.10 |
ENSMUST00000154668.1
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chrX_-_23266751 | 0.10 |
ENSMUST00000115316.2
|
Klhl13
|
kelch-like 13 |
| chr15_+_72913357 | 0.09 |
ENSMUST00000166418.2
|
Gm3150
|
predicted gene 3150 |
| chr5_-_48754521 | 0.09 |
ENSMUST00000101214.2
ENSMUST00000176191.1 |
Kcnip4
|
Kv channel interacting protein 4 |
| chr4_-_40757823 | 0.09 |
ENSMUST00000030117.4
|
Smu1
|
smu-1 suppressor of mec-8 and unc-52 homolog (C. elegans) |
| chr3_-_105052948 | 0.08 |
ENSMUST00000098763.2
|
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr8_+_9977707 | 0.08 |
ENSMUST00000139793.1
ENSMUST00000048216.5 |
Abhd13
|
abhydrolase domain containing 13 |
| chr4_-_121324912 | 0.07 |
ENSMUST00000097905.2
|
Gm12888
|
predicted gene 12888 |
| chr10_-_128804353 | 0.07 |
ENSMUST00000051011.7
|
Tmem198b
|
transmembrane protein 198b |
| chr17_+_42315947 | 0.07 |
ENSMUST00000048691.4
|
Ptchd4
|
patched domain containing 4 |
| chr13_-_21531084 | 0.07 |
ENSMUST00000045228.5
|
Zkscan8
|
zinc finger with KRAB and SCAN domains 8 |
| chr5_-_31220491 | 0.06 |
ENSMUST00000031032.7
|
Ppm1g
|
protein phosphatase 1G (formerly 2C), magnesium-dependent, gamma isoform |
| chr10_-_52195244 | 0.05 |
ENSMUST00000020045.3
|
Ros1
|
Ros1 proto-oncogene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmbox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 0.7 | 2.2 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.7 | 2.2 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.6 | 2.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.5 | 2.5 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 2.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.4 | 2.1 | GO:0031437 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.4 | 1.8 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.4 | 2.2 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.3 | 1.0 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.3 | 1.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.3 | 0.8 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.3 | 1.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.2 | 7.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 4.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 0.8 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.2 | 0.5 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 0.5 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.2 | 1.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 1.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 1.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 0.8 | GO:0035617 | stress granule disassembly(GO:0035617) plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 0.5 | GO:2000284 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 0.6 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 0.1 | 0.7 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.7 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 6.8 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.7 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 1.7 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 1.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.9 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 1.0 | GO:0007028 | cytoplasm organization(GO:0007028) negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 1.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 1.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 1.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.2 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 1.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.7 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 3.0 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.6 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.8 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 1.2 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 2.2 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 2.0 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.3 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 2.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 4.9 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.8 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.9 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.5 | 1.8 | GO:0036019 | endolysosome(GO:0036019) |
| 0.3 | 2.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.7 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 0.7 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.9 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 2.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.8 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.3 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.1 | 0.6 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 3.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 1.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 2.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 2.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 6.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.7 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 2.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 8.6 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.7 | 2.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.6 | 2.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.6 | 2.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.4 | 2.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 1.2 | GO:0008384 | NF-kappaB-inducing kinase activity(GO:0004704) IkappaB kinase activity(GO:0008384) |
| 0.2 | 1.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.2 | 1.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 2.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 0.8 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.8 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.2 | 0.5 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 1.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.8 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.6 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 2.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 1.4 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 1.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.3 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.4 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.7 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 2.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.5 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.1 | 0.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.9 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.0 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.2 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.1 | 1.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.7 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 1.2 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 1.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 3.3 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 2.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 2.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 4.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.0 | 1.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 2.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 2.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 4.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 0.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 2.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.7 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 1.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 2.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |