Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
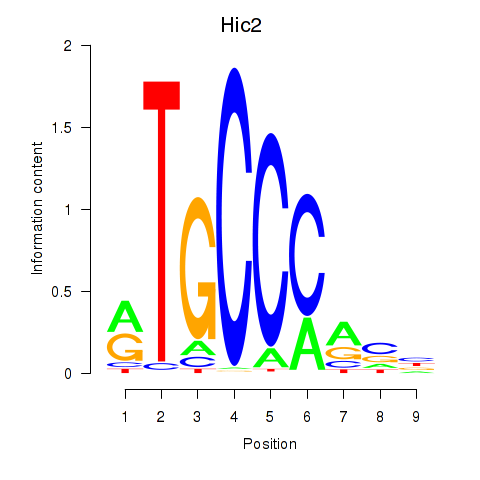
Results for Hic2
Z-value: 1.97
Transcription factors associated with Hic2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hic2
|
ENSMUSG00000050240.8 | hypermethylated in cancer 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hic2 | mm10_v2_chr16_+_17233560_17233664 | 0.68 | 5.7e-06 | Click! |
Activity profile of Hic2 motif
Sorted Z-values of Hic2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_109459843 | 18.92 |
ENSMUST00000173812.1
|
Dlk1
|
delta-like 1 homolog (Drosophila) |
| chr12_+_109549157 | 13.16 |
ENSMUST00000128458.1
ENSMUST00000150851.1 |
Meg3
|
maternally expressed 3 |
| chr11_+_7197780 | 9.98 |
ENSMUST00000020704.7
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr7_+_19411086 | 9.60 |
ENSMUST00000003643.1
|
Ckm
|
creatine kinase, muscle |
| chr11_+_87794206 | 9.49 |
ENSMUST00000121303.1
|
Mpo
|
myeloperoxidase |
| chr10_+_127725392 | 8.58 |
ENSMUST00000026466.3
|
Tac2
|
tachykinin 2 |
| chr11_+_115877497 | 8.39 |
ENSMUST00000144032.1
|
Myo15b
|
myosin XVB |
| chr11_-_102365111 | 8.35 |
ENSMUST00000006749.9
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr6_+_34412334 | 8.30 |
ENSMUST00000007449.8
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr14_+_51853699 | 8.15 |
ENSMUST00000169070.1
ENSMUST00000074477.6 |
Ear6
|
eosinophil-associated, ribonuclease A family, member 6 |
| chr18_+_34840575 | 7.95 |
ENSMUST00000043484.7
|
Reep2
|
receptor accessory protein 2 |
| chr12_+_109544498 | 7.88 |
ENSMUST00000126289.1
|
Meg3
|
maternally expressed 3 |
| chr12_+_109453455 | 7.65 |
ENSMUST00000109844.4
ENSMUST00000109842.2 ENSMUST00000109843.1 ENSMUST00000109846.4 ENSMUST00000173539.1 ENSMUST00000109841.2 |
Dlk1
|
delta-like 1 homolog (Drosophila) |
| chr4_+_130055010 | 7.43 |
ENSMUST00000123617.1
|
Col16a1
|
collagen, type XVI, alpha 1 |
| chr11_-_69617879 | 7.18 |
ENSMUST00000005334.2
|
Shbg
|
sex hormone binding globulin |
| chr10_-_128401218 | 7.10 |
ENSMUST00000042666.5
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr7_-_142578093 | 7.10 |
ENSMUST00000149974.1
ENSMUST00000152754.1 |
H19
|
H19 fetal liver mRNA |
| chr18_-_78123324 | 7.07 |
ENSMUST00000160292.1
ENSMUST00000091813.5 |
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr7_+_96211656 | 7.04 |
ENSMUST00000107165.1
|
Tenm4
|
teneurin transmembrane protein 4 |
| chr12_+_109452833 | 6.99 |
ENSMUST00000056110.8
|
Dlk1
|
delta-like 1 homolog (Drosophila) |
| chr7_-_142578139 | 6.89 |
ENSMUST00000136359.1
|
H19
|
H19 fetal liver mRNA |
| chr9_-_103480328 | 6.74 |
ENSMUST00000124310.2
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr7_-_24760311 | 6.58 |
ENSMUST00000063956.5
|
Cd177
|
CD177 antigen |
| chr11_+_115887601 | 6.49 |
ENSMUST00000167507.2
|
Myo15b
|
myosin XVB |
| chr14_-_20269162 | 6.46 |
ENSMUST00000024155.7
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr4_-_118457450 | 6.45 |
ENSMUST00000106375.1
ENSMUST00000006556.3 ENSMUST00000168404.1 |
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr19_+_10015016 | 6.41 |
ENSMUST00000137637.1
ENSMUST00000149967.1 |
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr2_-_163918683 | 6.39 |
ENSMUST00000044734.2
|
Rims4
|
regulating synaptic membrane exocytosis 4 |
| chr11_+_11684967 | 6.39 |
ENSMUST00000126058.1
ENSMUST00000141436.1 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr7_+_28540863 | 6.32 |
ENSMUST00000119180.2
|
Sycn
|
syncollin |
| chr7_-_126414855 | 6.23 |
ENSMUST00000032968.5
|
Cd19
|
CD19 antigen |
| chr11_+_4236411 | 6.15 |
ENSMUST00000075221.2
|
Osm
|
oncostatin M |
| chr12_+_111417430 | 6.09 |
ENSMUST00000072646.6
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr4_-_118457509 | 6.07 |
ENSMUST00000102671.3
|
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr8_-_10928449 | 6.07 |
ENSMUST00000040608.3
|
3930402G23Rik
|
RIKEN cDNA 3930402G23 gene |
| chr1_+_93006328 | 5.82 |
ENSMUST00000059676.4
|
Aqp12
|
aquaporin 12 |
| chr7_-_110982049 | 5.79 |
ENSMUST00000142368.1
|
Mrvi1
|
MRV integration site 1 |
| chr4_-_140774196 | 5.77 |
ENSMUST00000026381.6
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr9_+_58014990 | 5.63 |
ENSMUST00000034874.7
|
Cyp11a1
|
cytochrome P450, family 11, subfamily a, polypeptide 1 |
| chr12_+_102554966 | 5.51 |
ENSMUST00000021610.5
|
Chga
|
chromogranin A |
| chr17_+_25471564 | 5.50 |
ENSMUST00000025002.1
|
Tekt4
|
tektin 4 |
| chr7_+_142498832 | 5.49 |
ENSMUST00000078497.8
ENSMUST00000105953.3 ENSMUST00000179658.1 ENSMUST00000105954.3 ENSMUST00000105952.3 ENSMUST00000105955.1 ENSMUST00000074187.6 ENSMUST00000180152.1 ENSMUST00000105950.4 ENSMUST00000105957.3 ENSMUST00000169299.2 ENSMUST00000105958.3 ENSMUST00000105949.1 |
Tnnt3
|
troponin T3, skeletal, fast |
| chrX_+_93675088 | 5.32 |
ENSMUST00000045898.3
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr7_+_16906488 | 5.27 |
ENSMUST00000086101.5
ENSMUST00000144408.1 |
Ptgir
|
prostaglandin I receptor (IP) |
| chr17_-_57194170 | 5.23 |
ENSMUST00000005976.6
|
Tnfsf14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr6_+_29694204 | 5.23 |
ENSMUST00000046750.7
ENSMUST00000115250.3 |
Tspan33
|
tetraspanin 33 |
| chr3_+_68691424 | 5.21 |
ENSMUST00000107816.2
|
Il12a
|
interleukin 12a |
| chr17_-_35066170 | 4.96 |
ENSMUST00000174190.1
ENSMUST00000097337.1 |
AU023871
|
expressed sequence AU023871 |
| chr12_+_24831583 | 4.91 |
ENSMUST00000110942.3
ENSMUST00000078902.6 |
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr9_+_45138437 | 4.82 |
ENSMUST00000060125.5
|
Scn4b
|
sodium channel, type IV, beta |
| chr17_+_48247759 | 4.82 |
ENSMUST00000048065.5
|
Trem3
|
triggering receptor expressed on myeloid cells 3 |
| chr7_-_126704736 | 4.80 |
ENSMUST00000131415.1
|
Coro1a
|
coronin, actin binding protein 1A |
| chr12_-_79007276 | 4.80 |
ENSMUST00000056660.6
ENSMUST00000174721.1 |
Tmem229b
|
transmembrane protein 229B |
| chr2_-_170427828 | 4.78 |
ENSMUST00000013667.2
ENSMUST00000109152.2 ENSMUST00000068137.4 |
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr15_-_79285470 | 4.75 |
ENSMUST00000170955.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr11_+_98383811 | 4.72 |
ENSMUST00000008021.2
|
Tcap
|
titin-cap |
| chr8_+_23139064 | 4.71 |
ENSMUST00000033947.8
|
Ank1
|
ankyrin 1, erythroid |
| chr10_+_75571522 | 4.67 |
ENSMUST00000143226.1
ENSMUST00000124259.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr2_-_28563362 | 4.56 |
ENSMUST00000028161.5
|
Cel
|
carboxyl ester lipase |
| chr13_-_19619820 | 4.44 |
ENSMUST00000002885.6
|
Epdr1
|
ependymin related protein 1 (zebrafish) |
| chr10_-_19851459 | 4.43 |
ENSMUST00000059805.4
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr19_-_4615647 | 4.41 |
ENSMUST00000113822.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr11_-_120648104 | 4.40 |
ENSMUST00000026134.2
|
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr15_+_99224976 | 4.40 |
ENSMUST00000041415.3
|
Kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr15_-_79285502 | 4.38 |
ENSMUST00000165408.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr8_+_23139030 | 4.35 |
ENSMUST00000121075.1
|
Ank1
|
ankyrin 1, erythroid |
| chr4_-_64046925 | 4.33 |
ENSMUST00000107377.3
|
Tnc
|
tenascin C |
| chr15_+_76457438 | 4.23 |
ENSMUST00000043089.7
|
Scx
|
scleraxis |
| chr19_-_4877882 | 4.23 |
ENSMUST00000006626.3
|
Actn3
|
actinin alpha 3 |
| chr8_-_121907678 | 4.16 |
ENSMUST00000045557.9
|
Slc7a5
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
| chr1_-_75505641 | 4.16 |
ENSMUST00000155084.1
|
Obsl1
|
obscurin-like 1 |
| chr11_-_46312220 | 4.15 |
ENSMUST00000129474.1
ENSMUST00000093166.4 ENSMUST00000165599.2 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr7_+_110774240 | 4.09 |
ENSMUST00000147587.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr7_-_110982169 | 4.07 |
ENSMUST00000154466.1
|
Mrvi1
|
MRV integration site 1 |
| chr5_-_107723954 | 4.05 |
ENSMUST00000165344.1
|
Gfi1
|
growth factor independent 1 |
| chr7_+_142472080 | 4.05 |
ENSMUST00000105966.1
|
Lsp1
|
lymphocyte specific 1 |
| chr11_+_43528759 | 4.01 |
ENSMUST00000050574.6
|
Ccnjl
|
cyclin J-like |
| chr17_-_24527830 | 3.96 |
ENSMUST00000176353.1
ENSMUST00000176237.1 |
Traf7
|
TNF receptor-associated factor 7 |
| chr19_-_10203880 | 3.96 |
ENSMUST00000142241.1
ENSMUST00000116542.2 ENSMUST00000025651.5 ENSMUST00000156291.1 |
Fen1
|
flap structure specific endonuclease 1 |
| chr7_+_127211608 | 3.94 |
ENSMUST00000032910.6
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr11_+_32276400 | 3.91 |
ENSMUST00000020531.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr17_-_8101228 | 3.88 |
ENSMUST00000097422.4
|
Gm1604A
|
predicted gene 1604A |
| chr7_+_110768169 | 3.84 |
ENSMUST00000170374.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr12_+_109545390 | 3.82 |
ENSMUST00000146701.1
|
Meg3
|
maternally expressed 3 |
| chr15_+_57985873 | 3.79 |
ENSMUST00000050374.2
|
Fam83a
|
family with sequence similarity 83, member A |
| chr9_+_119052770 | 3.79 |
ENSMUST00000051386.6
ENSMUST00000074734.6 |
Vill
|
villin-like |
| chr13_+_21722057 | 3.71 |
ENSMUST00000110476.3
|
Hist1h2bm
|
histone cluster 1, H2bm |
| chr17_+_35821675 | 3.70 |
ENSMUST00000003635.6
|
Ier3
|
immediate early response 3 |
| chr9_+_8544196 | 3.70 |
ENSMUST00000050433.6
|
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr1_+_135132693 | 3.69 |
ENSMUST00000049449.4
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr8_+_105518736 | 3.68 |
ENSMUST00000034363.5
|
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr11_+_97050811 | 3.65 |
ENSMUST00000168565.1
|
Osbpl7
|
oxysterol binding protein-like 7 |
| chr3_+_108383829 | 3.63 |
ENSMUST00000090561.3
ENSMUST00000102629.1 ENSMUST00000128089.1 |
Psrc1
|
proline/serine-rich coiled-coil 1 |
| chr7_-_135716374 | 3.62 |
ENSMUST00000033310.7
|
Mki67
|
antigen identified by monoclonal antibody Ki 67 |
| chr8_-_46211284 | 3.62 |
ENSMUST00000034049.4
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chr9_-_37552904 | 3.60 |
ENSMUST00000065668.5
|
Nrgn
|
neurogranin |
| chr9_+_119052863 | 3.59 |
ENSMUST00000131647.1
|
Vill
|
villin-like |
| chr7_+_19577287 | 3.57 |
ENSMUST00000108453.1
|
Zfp296
|
zinc finger protein 296 |
| chr7_+_80294450 | 3.55 |
ENSMUST00000163812.2
ENSMUST00000047558.7 ENSMUST00000174199.1 ENSMUST00000173824.1 ENSMUST00000174172.1 |
Prc1
|
protein regulator of cytokinesis 1 |
| chr6_-_78378851 | 3.52 |
ENSMUST00000089667.1
ENSMUST00000167492.1 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr19_-_4615453 | 3.51 |
ENSMUST00000053597.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr11_+_115154139 | 3.50 |
ENSMUST00000021076.5
|
Rab37
|
RAB37, member of RAS oncogene family |
| chr17_-_25952565 | 3.48 |
ENSMUST00000162431.1
|
A930017K11Rik
|
RIKEN cDNA A930017K11 gene |
| chr14_+_118854695 | 3.47 |
ENSMUST00000100314.3
|
Cldn10
|
claudin 10 |
| chr11_+_97050594 | 3.45 |
ENSMUST00000090020.5
|
Osbpl7
|
oxysterol binding protein-like 7 |
| chrX_-_49788204 | 3.44 |
ENSMUST00000114893.1
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr4_-_108383349 | 3.44 |
ENSMUST00000053157.6
|
Fam159a
|
family with sequence similarity 159, member A |
| chr7_-_126704816 | 3.41 |
ENSMUST00000032949.7
|
Coro1a
|
coronin, actin binding protein 1A |
| chr2_-_181156993 | 3.40 |
ENSMUST00000055990.7
|
Eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr1_+_134182150 | 3.39 |
ENSMUST00000156873.1
|
Chi3l1
|
chitinase 3-like 1 |
| chr11_+_76998595 | 3.37 |
ENSMUST00000108402.2
ENSMUST00000021195.4 |
Slc6a4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
| chr3_+_88607742 | 3.37 |
ENSMUST00000175903.1
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr7_-_110982443 | 3.37 |
ENSMUST00000005751.6
|
Mrvi1
|
MRV integration site 1 |
| chr5_-_53707532 | 3.35 |
ENSMUST00000031093.3
|
Cckar
|
cholecystokinin A receptor |
| chr17_-_24527925 | 3.35 |
ENSMUST00000176652.1
|
Traf7
|
TNF receptor-associated factor 7 |
| chr19_+_4154606 | 3.29 |
ENSMUST00000061086.8
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr7_+_110773658 | 3.25 |
ENSMUST00000143786.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr5_-_112392213 | 3.24 |
ENSMUST00000031291.7
|
Asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr14_-_51057242 | 3.19 |
ENSMUST00000089798.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr7_-_67803489 | 3.18 |
ENSMUST00000181235.1
|
4833412C05Rik
|
RIKEN cDNA 4833412C05 gene |
| chr19_-_34255325 | 3.14 |
ENSMUST00000039631.8
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr10_+_79930419 | 3.11 |
ENSMUST00000131118.1
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr12_-_113422730 | 3.10 |
ENSMUST00000177715.1
ENSMUST00000103426.1 |
Ighm
|
immunoglobulin heavy constant mu |
| chr17_+_28207778 | 3.08 |
ENSMUST00000002327.5
|
Def6
|
differentially expressed in FDCP 6 |
| chr9_+_107906866 | 3.07 |
ENSMUST00000035203.7
|
Mst1r
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr4_-_117929466 | 3.06 |
ENSMUST00000097913.2
|
Artn
|
artemin |
| chr3_-_98893209 | 3.05 |
ENSMUST00000029464.7
|
Hao2
|
hydroxyacid oxidase 2 |
| chr4_-_129440800 | 3.04 |
ENSMUST00000053042.5
ENSMUST00000106046.1 |
Zbtb8b
|
zinc finger and BTB domain containing 8b |
| chr1_+_134193432 | 3.04 |
ENSMUST00000038445.6
|
Mybph
|
myosin binding protein H |
| chr15_-_55090422 | 3.00 |
ENSMUST00000110231.1
ENSMUST00000023059.6 |
Dscc1
|
defective in sister chromatid cohesion 1 homolog (S. cerevisiae) |
| chrX_-_104671048 | 2.98 |
ENSMUST00000042070.5
|
Zdhhc15
|
zinc finger, DHHC domain containing 15 |
| chr1_+_174041933 | 2.98 |
ENSMUST00000052975.4
|
Olfr433
|
olfactory receptor 433 |
| chr12_+_85599388 | 2.97 |
ENSMUST00000050687.6
|
Jdp2
|
Jun dimerization protein 2 |
| chr11_-_12026237 | 2.94 |
ENSMUST00000150972.1
|
Grb10
|
growth factor receptor bound protein 10 |
| chr13_-_70841790 | 2.93 |
ENSMUST00000080145.6
ENSMUST00000109694.2 |
Adamts16
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 16 |
| chr7_-_103813913 | 2.92 |
ENSMUST00000098192.3
|
Hbb-bt
|
hemoglobin, beta adult t chain |
| chr7_+_43440782 | 2.92 |
ENSMUST00000040227.1
|
Cldnd2
|
claudin domain containing 2 |
| chr17_-_56830916 | 2.91 |
ENSMUST00000002444.7
ENSMUST00000086801.5 |
Rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr11_-_12026732 | 2.90 |
ENSMUST00000143915.1
|
Grb10
|
growth factor receptor bound protein 10 |
| chrX_-_8145679 | 2.89 |
ENSMUST00000115619.1
ENSMUST00000115617.3 ENSMUST00000040010.3 |
Rbm3
|
RNA binding motif protein 3 |
| chr11_-_106349389 | 2.89 |
ENSMUST00000021056.7
|
Scn4a
|
sodium channel, voltage-gated, type IV, alpha |
| chr11_-_69602741 | 2.88 |
ENSMUST00000138694.1
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr7_-_100856289 | 2.86 |
ENSMUST00000139604.1
|
Relt
|
RELT tumor necrosis factor receptor |
| chr11_+_69095217 | 2.84 |
ENSMUST00000101004.2
|
Per1
|
period circadian clock 1 |
| chr11_+_115974930 | 2.82 |
ENSMUST00000106460.2
|
Itgb4
|
integrin beta 4 |
| chrX_+_93654863 | 2.82 |
ENSMUST00000113933.2
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr2_+_29869484 | 2.80 |
ENSMUST00000047521.6
ENSMUST00000134152.1 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr8_+_12915879 | 2.77 |
ENSMUST00000110876.2
ENSMUST00000110879.2 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr6_+_128399766 | 2.77 |
ENSMUST00000001561.5
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr3_+_84666192 | 2.76 |
ENSMUST00000107682.1
|
Tmem154
|
transmembrane protein 154 |
| chr3_+_123446913 | 2.76 |
ENSMUST00000029603.8
|
Prss12
|
protease, serine, 12 neurotrypsin (motopsin) |
| chr14_+_57524734 | 2.74 |
ENSMUST00000089494.4
|
Il17d
|
interleukin 17D |
| chr15_-_101712891 | 2.74 |
ENSMUST00000023709.5
|
Krt5
|
keratin 5 |
| chr2_-_165473187 | 2.72 |
ENSMUST00000029208.8
ENSMUST00000109279.2 |
Slc13a3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr9_-_107985863 | 2.71 |
ENSMUST00000048568.4
|
Fam212a
|
family with sequence similarity 212, member A |
| chr2_-_170194033 | 2.71 |
ENSMUST00000180625.1
|
Gm17619
|
predicted gene, 17619 |
| chr2_-_92370968 | 2.71 |
ENSMUST00000176774.1
|
Gyltl1b
|
glycosyltransferase-like 1B |
| chr6_-_29212240 | 2.71 |
ENSMUST00000160878.1
ENSMUST00000078155.5 |
Impdh1
|
inosine 5'-phosphate dehydrogenase 1 |
| chr9_-_119578981 | 2.70 |
ENSMUST00000117911.1
ENSMUST00000120420.1 |
Scn5a
|
sodium channel, voltage-gated, type V, alpha |
| chrX_-_51205990 | 2.67 |
ENSMUST00000114876.2
|
Mbnl3
|
muscleblind-like 3 (Drosophila) |
| chr1_-_75506331 | 2.64 |
ENSMUST00000113567.2
ENSMUST00000113565.2 |
Obsl1
|
obscurin-like 1 |
| chr7_+_28982832 | 2.63 |
ENSMUST00000085835.6
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr17_-_32403551 | 2.62 |
ENSMUST00000135618.1
ENSMUST00000063824.7 |
Rasal3
|
RAS protein activator like 3 |
| chr4_-_127330799 | 2.61 |
ENSMUST00000046532.3
|
Gjb3
|
gap junction protein, beta 3 |
| chr10_-_61273242 | 2.61 |
ENSMUST00000120336.1
|
Adamts14
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 14 |
| chr6_+_128399881 | 2.60 |
ENSMUST00000120405.1
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr13_-_3804307 | 2.59 |
ENSMUST00000077698.3
|
Calml3
|
calmodulin-like 3 |
| chr2_+_130277157 | 2.58 |
ENSMUST00000028890.8
ENSMUST00000159373.1 |
Nop56
|
NOP56 ribonucleoprotein |
| chr2_+_92915080 | 2.56 |
ENSMUST00000028648.2
|
Syt13
|
synaptotagmin XIII |
| chr4_+_154960915 | 2.56 |
ENSMUST00000049621.6
|
Hes5
|
hairy and enhancer of split 5 (Drosophila) |
| chr14_+_60378242 | 2.53 |
ENSMUST00000022561.6
|
Amer2
|
APC membrane recruitment 2 |
| chr2_-_33887862 | 2.53 |
ENSMUST00000041555.3
|
Mvb12b
|
multivesicular body subunit 12B |
| chr2_-_29869785 | 2.52 |
ENSMUST00000047607.1
|
2600006K01Rik
|
RIKEN cDNA 2600006K01 gene |
| chr17_-_32403526 | 2.51 |
ENSMUST00000137458.1
|
Rasal3
|
RAS protein activator like 3 |
| chr3_-_153725062 | 2.51 |
ENSMUST00000064460.5
|
St6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr15_-_66831625 | 2.49 |
ENSMUST00000164163.1
|
Sla
|
src-like adaptor |
| chrX_+_74329058 | 2.48 |
ENSMUST00000004326.3
|
Plxna3
|
plexin A3 |
| chr4_-_152477433 | 2.46 |
ENSMUST00000159186.1
ENSMUST00000162017.1 ENSMUST00000030768.2 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr2_-_92370999 | 2.45 |
ENSMUST00000176810.1
ENSMUST00000090582.4 |
Gyltl1b
|
glycosyltransferase-like 1B |
| chr11_+_96931387 | 2.45 |
ENSMUST00000107633.1
|
Prr15l
|
proline rich 15-like |
| chr17_-_25433775 | 2.44 |
ENSMUST00000159610.1
ENSMUST00000159048.1 ENSMUST00000078496.5 |
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr11_-_102897123 | 2.44 |
ENSMUST00000067444.3
|
Gfap
|
glial fibrillary acidic protein |
| chr2_-_166713758 | 2.44 |
ENSMUST00000036719.5
|
Prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr11_+_87755567 | 2.43 |
ENSMUST00000123700.1
|
A430104N18Rik
|
RIKEN cDNA A430104N18 gene |
| chr13_+_20090538 | 2.42 |
ENSMUST00000072519.5
|
Elmo1
|
engulfment and cell motility 1 |
| chr5_+_30666886 | 2.42 |
ENSMUST00000144742.1
|
Cenpa
|
centromere protein A |
| chr2_+_106695594 | 2.42 |
ENSMUST00000016530.7
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr17_-_48409729 | 2.38 |
ENSMUST00000160319.1
ENSMUST00000159535.1 ENSMUST00000078800.6 ENSMUST00000046719.7 ENSMUST00000162460.1 |
Nfya
|
nuclear transcription factor-Y alpha |
| chr4_+_133240778 | 2.36 |
ENSMUST00000030677.6
|
Map3k6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr8_-_85380964 | 2.36 |
ENSMUST00000122452.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr15_-_98607611 | 2.36 |
ENSMUST00000096224.4
|
Adcy6
|
adenylate cyclase 6 |
| chr4_-_134128707 | 2.35 |
ENSMUST00000105879.1
ENSMUST00000030651.8 |
Sh3bgrl3
|
SH3 domain binding glutamic acid-rich protein-like 3 |
| chr19_-_46044914 | 2.32 |
ENSMUST00000026252.7
|
Ldb1
|
LIM domain binding 1 |
| chr18_+_62180119 | 2.32 |
ENSMUST00000067743.1
|
Gm9949
|
predicted gene 9949 |
| chr10_-_80421847 | 2.32 |
ENSMUST00000156244.1
|
Tcf3
|
transcription factor 3 |
| chr4_+_140906344 | 2.31 |
ENSMUST00000030765.6
|
Padi2
|
peptidyl arginine deiminase, type II |
| chrX_+_134308084 | 2.30 |
ENSMUST00000081064.5
ENSMUST00000101251.1 ENSMUST00000129782.1 |
Cenpi
|
centromere protein I |
| chr12_+_110279228 | 2.30 |
ENSMUST00000097228.4
|
Dio3
|
deiodinase, iodothyronine type III |
| chr18_-_62179948 | 2.30 |
ENSMUST00000053640.3
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr4_+_136284708 | 2.29 |
ENSMUST00000130223.1
|
Zfp46
|
zinc finger protein 46 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hic2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 11.9 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 3.2 | 9.5 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 2.4 | 7.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 2.4 | 7.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 2.1 | 6.4 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 1.8 | 5.5 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 1.8 | 9.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 1.8 | 7.0 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 1.7 | 5.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 1.7 | 5.2 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 1.7 | 8.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 1.7 | 10.2 | GO:0032796 | uropod organization(GO:0032796) |
| 1.5 | 4.4 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 1.5 | 5.9 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 1.3 | 11.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.2 | 3.7 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 1.2 | 6.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 1.2 | 3.6 | GO:0015866 | ADP transport(GO:0015866) |
| 1.2 | 11.5 | GO:0051133 | regulation of NK T cell activation(GO:0051133) |
| 1.2 | 3.5 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 1.1 | 8.0 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 1.1 | 3.4 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 1.1 | 5.5 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 1.1 | 4.4 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.1 | 4.3 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 1.1 | 14.0 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 1.1 | 1.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 1.0 | 3.1 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.0 | 30.9 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 1.0 | 4.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 1.0 | 1.0 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 1.0 | 5.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.0 | 1.0 | GO:0060983 | epicardium-derived cardiac vascular smooth muscle cell differentiation(GO:0060983) |
| 1.0 | 2.9 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 1.0 | 2.9 | GO:0046144 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.9 | 12.8 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.9 | 14.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.9 | 0.9 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.9 | 0.9 | GO:0042223 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.9 | 6.1 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.8 | 4.2 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.8 | 3.4 | GO:0060032 | notochord regression(GO:0060032) |
| 0.8 | 3.4 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.8 | 24.5 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.8 | 1.6 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) positive regulation of dermatome development(GO:0061184) |
| 0.8 | 5.5 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 0.7 | 2.2 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.7 | 2.2 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.7 | 2.2 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.7 | 2.9 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.7 | 7.1 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.7 | 2.7 | GO:0015744 | succinate transport(GO:0015744) |
| 0.7 | 3.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.7 | 4.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.7 | 2.0 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.7 | 2.0 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.6 | 1.9 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.6 | 0.6 | GO:1902943 | regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) |
| 0.6 | 3.2 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.6 | 3.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.6 | 2.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.6 | 3.0 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.6 | 7.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.6 | 2.4 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.6 | 4.7 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.6 | 1.7 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.6 | 2.3 | GO:1904504 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.6 | 2.8 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.6 | 2.8 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.6 | 2.8 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.6 | 1.7 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.6 | 2.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.6 | 1.7 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.5 | 4.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.5 | 2.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.5 | 1.6 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.5 | 1.6 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.5 | 1.6 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.5 | 5.8 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.5 | 2.0 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.5 | 1.5 | GO:0060220 | camera-type eye photoreceptor cell fate commitment(GO:0060220) negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 0.5 | 1.0 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.5 | 1.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.5 | 2.0 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.5 | 1.5 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.5 | 1.5 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.5 | 2.0 | GO:0021586 | pons maturation(GO:0021586) |
| 0.5 | 1.0 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.5 | 3.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.5 | 1.9 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.5 | 1.4 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.5 | 0.9 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.5 | 1.4 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.4 | 2.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.4 | 1.8 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.4 | 1.3 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.4 | 3.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.4 | 4.6 | GO:0035878 | nail development(GO:0035878) |
| 0.4 | 2.1 | GO:0035993 | subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) superior vena cava morphogenesis(GO:0060578) |
| 0.4 | 2.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.4 | 1.6 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.4 | 3.6 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.4 | 2.0 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.4 | 0.8 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.4 | 6.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.4 | 1.6 | GO:0061743 | motor learning(GO:0061743) |
| 0.4 | 9.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.4 | 1.6 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.4 | 9.8 | GO:0033622 | integrin activation(GO:0033622) |
| 0.4 | 2.0 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.4 | 1.2 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.4 | 0.4 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.4 | 2.3 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.4 | 2.3 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.4 | 1.1 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.4 | 0.7 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.4 | 1.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.4 | 1.8 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.4 | 5.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.4 | 4.6 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 2.1 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.3 | 0.7 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 0.3 | 1.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.3 | 2.0 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.3 | 0.7 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.3 | 1.0 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.3 | 1.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.3 | 3.5 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.3 | 8.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 1.3 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.3 | 5.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 5.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.3 | 5.3 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.3 | 1.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.3 | 2.7 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.3 | 3.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.3 | 0.3 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.3 | 1.8 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.3 | 1.5 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.3 | 1.5 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.3 | 4.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 1.8 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.3 | 5.5 | GO:0099625 | ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.3 | 0.6 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.3 | 0.6 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.3 | 2.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.3 | 2.6 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.3 | 0.9 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.3 | 2.5 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) |
| 0.3 | 0.8 | GO:0048866 | stem cell fate specification(GO:0048866) |
| 0.3 | 0.8 | GO:0072708 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 0.3 | 10.1 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.3 | 2.2 | GO:0034651 | cortisol metabolic process(GO:0034650) cortisol biosynthetic process(GO:0034651) positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 1.4 | GO:0032899 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) regulation of neurotrophin production(GO:0032899) positive regulation of neurotrophin production(GO:0032901) |
| 0.3 | 1.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.3 | 3.5 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.3 | 5.1 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.3 | 0.8 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.3 | 5.8 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.3 | 2.4 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 1.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.3 | 3.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.3 | 1.0 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.2 | 1.0 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.2 | 0.7 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.2 | 0.7 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 1.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 4.6 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 0.5 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.2 | 1.2 | GO:0042908 | xenobiotic transport(GO:0042908) dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 1.2 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.2 | 1.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 4.7 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.2 | 2.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 1.6 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.2 | 2.3 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.2 | 0.5 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.2 | 1.8 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.2 | 0.7 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.2 | 0.9 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) regulation of PERK-mediated unfolded protein response(GO:1903897) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.2 | 0.7 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.2 | 0.9 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.2 | 1.3 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.2 | 3.7 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.2 | 0.9 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.2 | 3.0 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.2 | 1.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 1.5 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 0.6 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.2 | 0.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 | 3.9 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.2 | 4.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 6.3 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.2 | 1.2 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.2 | 1.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.4 | GO:1904996 | regulation of leukocyte adhesion to vascular endothelial cell(GO:1904994) positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.2 | 1.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 | 1.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 0.4 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.2 | 0.7 | GO:0009816 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.2 | 0.7 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 0.9 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.2 | 1.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 0.5 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 1.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.2 | 0.5 | GO:0099542 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.2 | 1.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 8.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.2 | 0.5 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.2 | 0.5 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.2 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 | 0.8 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.2 | 4.7 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.2 | 0.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 2.4 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.2 | 0.8 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 1.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 1.9 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.2 | 1.3 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.2 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 1.4 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.1 | 2.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.7 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 1.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 8.0 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.1 | 1.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.4 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.7 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.6 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.1 | 0.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 6.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.4 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.8 | GO:0006172 | ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.1 | 3.1 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.1 | 0.5 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.5 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 | 0.4 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 1.4 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.6 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 2.7 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.1 | 1.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 1.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 2.5 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.7 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 1.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 1.2 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 1.7 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 1.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 5.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 6.4 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 2.1 | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.1 | 4.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.4 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 1.9 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 1.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 4.9 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 0.7 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 1.4 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.1 | 4.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.7 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 1.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 1.2 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.8 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.9 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.8 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.9 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.8 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.1 | 0.5 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.3 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.1 | 1.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.5 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.9 | GO:0061626 | neural crest cell migration involved in heart formation(GO:0003147) pharyngeal arch artery morphogenesis(GO:0061626) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 0.8 | GO:0042482 | positive regulation of odontogenesis(GO:0042482) |
| 0.1 | 1.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 1.0 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 0.6 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 1.6 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 1.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.6 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 3.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 1.0 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 2.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 2.3 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 0.7 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.2 | GO:1903978 | regulation of microglial cell activation(GO:1903978) |
| 0.1 | 0.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.8 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 2.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.2 | GO:0061309 | cardiac neural crest cell development involved in outflow tract morphogenesis(GO:0061309) |
| 0.1 | 0.2 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.1 | 0.8 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 2.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.4 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.7 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.9 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.1 | 0.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 3.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.7 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.1 | 0.6 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.3 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.1 | 0.5 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.8 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 1.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.7 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 0.2 | GO:0080144 | amino acid homeostasis(GO:0080144) |
| 0.1 | 0.9 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 2.2 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.1 | 1.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.2 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 0.7 | GO:0043267 | negative regulation of potassium ion transport(GO:0043267) |
| 0.1 | 1.9 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 1.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.4 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 | 1.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 5.0 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 0.8 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.1 | 2.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.3 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.3 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.5 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.6 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.3 | GO:0035617 | stress granule disassembly(GO:0035617) plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 1.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 1.2 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.1 | 0.9 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 1.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 1.7 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 3.1 | GO:0060998 | regulation of dendritic spine development(GO:0060998) |
| 0.0 | 1.0 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.1 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.0 | 0.7 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.7 | GO:2001259 | positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.5 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.0 | 0.8 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 1.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 1.1 | GO:0071384 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.7 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 1.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0009397 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0050881 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 0.1 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 2.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 2.7 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.3 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 1.0 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.4 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0046015 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) carbon catabolite regulation of transcription(GO:0045990) carbon catabolite activation of transcription(GO:0045991) regulation of transcription by glucose(GO:0046015) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0021634 | optic nerve formation(GO:0021634) regulation of metanephros size(GO:0035566) optic chiasma development(GO:0061360) positive regulation of metanephric glomerulus development(GO:0072300) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.0 | 1.5 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.5 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.6 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.2 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.6 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0044819 | mitotic G1 DNA damage checkpoint(GO:0031571) mitotic G1/S transition checkpoint(GO:0044819) |
| 0.0 | 0.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.0 | 1.0 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.5 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.2 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 1.1 | 5.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.9 | 6.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.8 | 2.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.8 | 9.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.7 | 10.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.7 | 2.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.6 | 2.4 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.6 | 15.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.6 | 3.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.6 | 6.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.5 | 2.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.5 | 8.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.5 | 3.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.5 | 2.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.5 | 1.0 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.5 | 1.4 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.4 | 3.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.4 | 2.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.4 | 10.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.4 | 6.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.4 | 2.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.4 | 1.5 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 2.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 10.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 6.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.4 | 1.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.3 | 1.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.3 | 3.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 3.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.3 | 3.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 1.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.3 | 5.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.3 | 1.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.3 | 5.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 1.5 | GO:0071914 | prominosome(GO:0071914) |
| 0.3 | 1.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 3.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 4.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 6.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 0.7 | GO:0005595 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 0.2 | 6.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 3.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 4.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 1.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 1.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 3.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 5.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 1.0 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 2.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 0.8 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.2 | 2.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 14.7 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 0.7 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.2 | 0.9 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.2 | 13.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.2 | 1.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 3.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 1.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 5.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 0.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 2.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 4.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 1.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 10.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.8 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 4.7 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 4.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.9 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 3.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 2.0 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 5.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 2.6 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 2.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.6 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 2.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.2 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 1.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 2.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.2 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 37.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 4.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 6.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 8.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 6.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 4.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 27.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 3.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 10.4 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 1.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 1.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.8 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 2.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.4 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 0.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.2 | GO:0016507 | mitochondrial fatty acid beta-oxidation multienzyme complex(GO:0016507) fatty acid beta-oxidation multienzyme complex(GO:0036125) |
| 0.1 | 1.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 6.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.8 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 1.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 3.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 4.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 2.6 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 2.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 4.0 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 3.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 1.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.3 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 2.2 | GO:0070401 | NADP+ binding(GO:0070401) |
| 1.8 | 9.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.8 | 5.3 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 1.7 | 5.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 1.6 | 8.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.6 | 9.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.5 | 4.6 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 1.3 | 5.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 1.3 | 11.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.2 | 3.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.2 | 7.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 1.1 | 10.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 1.1 | 3.4 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 1.1 | 7.5 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.0 | 10.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.0 | 5.9 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.0 | 3.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.9 | 4.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.9 | 5.5 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.9 | 2.7 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.8 | 3.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.8 | 4.0 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.8 | 3.9 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.8 | 2.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.8 | 3.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.8 | 4.6 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.7 | 2.9 | GO:0031720 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 0.7 | 2.9 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.7 | 4.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.7 | 2.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.7 | 2.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.6 | 1.9 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.6 | 2.5 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.6 | 5.7 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.6 | 2.5 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.6 | 4.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.6 | 4.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 5.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.5 | 4.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 5.4 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.5 | 1.6 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.5 | 9.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.5 | 9.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.5 | 1.5 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.5 | 4.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.5 | 1.9 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.5 | 4.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.5 | 2.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.4 | 8.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 0.9 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.4 | 1.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 1.7 | GO:0002046 | opsin binding(GO:0002046) |
| 0.4 | 3.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 1.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.4 | 2.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.4 | 2.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.4 | 1.6 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.4 | 4.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 3.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.4 | 1.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.4 | 1.5 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.4 | 1.9 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.4 | 1.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.4 | 1.5 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.3 | 1.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 1.7 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.3 | 2.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 5.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 1.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 2.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 1.6 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 2.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 2.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.3 | 3.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 2.8 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.3 | 0.9 | GO:0019002 | GMP binding(GO:0019002) |
| 0.3 | 0.9 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.3 | 1.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.3 | 3.6 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 3.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 2.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.3 | 1.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 2.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 0.8 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.3 | 1.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.3 | 2.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 3.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 1.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 1.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.3 | 1.8 | GO:0019864 | IgG binding(GO:0019864) |
| 0.3 | 5.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 0.7 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.2 | 5.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 1.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 1.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 0.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 1.4 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.2 | 4.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 4.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 0.7 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 0.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 4.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 2.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 2.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 1.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 6.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 2.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.8 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 1.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 6.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.2 | 3.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.2 | 1.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 5.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 6.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 1.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 0.8 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 0.6 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.2 | 8.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 1.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 0.5 | GO:0070122 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 0.2 | 0.5 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.2 | 1.0 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.2 | 1.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 2.5 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.2 | 2.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 0.5 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.2 | 0.5 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 7.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 1.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 0.6 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 1.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.0 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.8 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.8 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.7 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.8 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.8 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 1.8 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 1.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.9 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 2.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.0 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 0.7 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 3.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.2 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.1 | 1.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 2.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 2.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.5 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 1.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 7.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.4 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 2.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 2.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.5 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.1 | 2.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 1.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 4.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 4.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.6 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.9 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 2.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 7.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 4.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.6 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 1.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.7 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 3.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 7.7 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 0.5 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.1 | 0.6 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 4.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.4 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 1.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 39.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 2.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 7.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 2.9 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 1.0 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 1.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.6 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 5.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 1.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 3.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 3.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 1.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 1.7 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 1.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 4.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.2 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 39.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.5 | 6.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 2.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.3 | 4.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.3 | 6.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 10.9 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.3 | 3.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 9.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 3.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 8.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 7.5 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.2 | 8.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 5.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 5.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 2.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.2 | 6.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 9.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 2.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 11.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 7.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 6.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 6.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 3.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 8.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 4.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 4.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 0.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 2.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 0.7 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 0.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.2 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 1.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 1.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 2.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 4.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 33.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.6 | 1.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.5 | 11.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.4 | 5.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.4 | 4.8 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.4 | 14.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 14.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 3.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.3 | 8.6 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.3 | 2.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.3 | 5.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 4.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.3 | 14.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 5.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 5.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 7.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 7.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 11.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 4.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 14.0 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.2 | 3.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 9.3 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.2 | 2.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 2.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 4.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 3.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 8.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 1.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 5.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 3.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 2.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 6.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.8 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 5.9 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 4.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 7.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 4.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 3.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 4.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 7.9 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.7 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 8.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 1.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 8.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 0.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 0.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 1.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 2.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 4.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME OPIOID SIGNALLING | Genes involved in Opioid Signalling |
| 0.0 | 1.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 2.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |