Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hey2
Z-value: 0.86
Transcription factors associated with Hey2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hey2
|
ENSMUSG00000019789.8 | hairy/enhancer-of-split related with YRPW motif 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hey2 | mm10_v2_chr10_-_30842765_30842801 | -0.07 | 7.0e-01 | Click! |
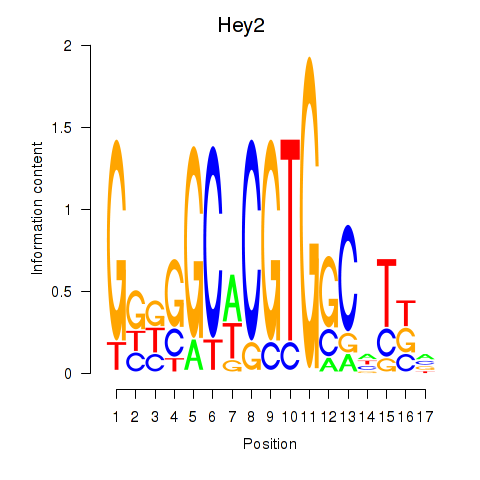
Activity profile of Hey2 motif
Sorted Z-values of Hey2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_72824482 | 5.99 |
ENSMUST00000047328.4
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr13_+_58807884 | 3.95 |
ENSMUST00000079828.5
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr8_+_105269837 | 3.69 |
ENSMUST00000172525.1
ENSMUST00000174837.1 ENSMUST00000173859.1 |
Hsf4
|
heat shock transcription factor 4 |
| chr8_+_105269788 | 3.63 |
ENSMUST00000036127.2
ENSMUST00000163734.2 |
Hsf4
|
heat shock transcription factor 4 |
| chr7_-_25658726 | 3.27 |
ENSMUST00000071329.6
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr18_-_61911783 | 2.78 |
ENSMUST00000049378.8
ENSMUST00000166783.1 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr4_-_57300751 | 2.69 |
ENSMUST00000151964.1
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr4_+_116877376 | 2.55 |
ENSMUST00000044823.3
|
Zswim5
|
zinc finger SWIM-type containing 5 |
| chr5_+_35757875 | 2.44 |
ENSMUST00000101280.3
ENSMUST00000054598.5 ENSMUST00000114205.1 ENSMUST00000114206.2 |
Ablim2
|
actin-binding LIM protein 2 |
| chr10_+_122448499 | 2.18 |
ENSMUST00000020323.5
|
Avpr1a
|
arginine vasopressin receptor 1A |
| chr1_-_166309585 | 2.15 |
ENSMUST00000168347.1
|
5330438I03Rik
|
RIKEN cDNA 5330438I03 gene |
| chr5_+_35757951 | 2.10 |
ENSMUST00000114204.1
ENSMUST00000129347.1 |
Ablim2
|
actin-binding LIM protein 2 |
| chr5_+_141241490 | 2.01 |
ENSMUST00000085774.4
|
Sdk1
|
sidekick homolog 1 (chicken) |
| chr1_+_87264345 | 1.95 |
ENSMUST00000118687.1
ENSMUST00000027472.6 |
Efhd1
|
EF hand domain containing 1 |
| chr5_-_25100624 | 1.93 |
ENSMUST00000030784.7
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr1_+_82586942 | 1.90 |
ENSMUST00000113457.2
|
Col4a3
|
collagen, type IV, alpha 3 |
| chr8_+_119446719 | 1.76 |
ENSMUST00000098363.3
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr14_-_70351354 | 1.74 |
ENSMUST00000143153.1
ENSMUST00000127000.1 ENSMUST00000068044.7 ENSMUST00000022688.3 |
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr7_+_30169861 | 1.70 |
ENSMUST00000085668.4
|
Gm5113
|
predicted gene 5113 |
| chr15_+_55112420 | 1.69 |
ENSMUST00000100660.4
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr9_+_57697612 | 1.66 |
ENSMUST00000034865.4
|
Cyp1a1
|
cytochrome P450, family 1, subfamily a, polypeptide 1 |
| chr15_+_55112317 | 1.63 |
ENSMUST00000096433.3
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr1_-_82586781 | 1.58 |
ENSMUST00000087050.5
|
Col4a4
|
collagen, type IV, alpha 4 |
| chr3_+_81932601 | 1.50 |
ENSMUST00000029649.2
|
Ctso
|
cathepsin O |
| chr15_+_80671829 | 1.48 |
ENSMUST00000023044.5
|
Fam83f
|
family with sequence similarity 83, member F |
| chr1_+_90915064 | 1.43 |
ENSMUST00000027528.6
|
Mlph
|
melanophilin |
| chr11_+_70092634 | 1.42 |
ENSMUST00000102572.1
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chrX_-_52165252 | 1.41 |
ENSMUST00000033450.2
|
Gpc4
|
glypican 4 |
| chr2_+_140395446 | 1.40 |
ENSMUST00000110061.1
|
Macrod2
|
MACRO domain containing 2 |
| chr2_-_130424242 | 1.39 |
ENSMUST00000089581.4
|
Pced1a
|
PC-esterase domain containing 1A |
| chr5_-_138996087 | 1.38 |
ENSMUST00000110897.1
|
Pdgfa
|
platelet derived growth factor, alpha |
| chr11_+_70092705 | 1.35 |
ENSMUST00000124721.1
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr15_+_57694651 | 1.35 |
ENSMUST00000096430.4
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr2_-_168741898 | 1.33 |
ENSMUST00000109176.1
ENSMUST00000178504.1 |
Atp9a
|
ATPase, class II, type 9A |
| chr11_+_70092653 | 1.32 |
ENSMUST00000143772.1
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr10_-_78487842 | 1.26 |
ENSMUST00000069431.4
|
Gm9978
|
predicted gene 9978 |
| chr7_+_44384604 | 1.23 |
ENSMUST00000130707.1
ENSMUST00000130844.1 |
Syt3
|
synaptotagmin III |
| chr8_+_85060055 | 1.23 |
ENSMUST00000095220.3
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr17_-_45686120 | 1.22 |
ENSMUST00000143907.1
ENSMUST00000127065.1 |
Tmem63b
|
transmembrane protein 63b |
| chr5_+_33104219 | 1.18 |
ENSMUST00000011178.2
|
Slc5a1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr7_+_101321703 | 1.15 |
ENSMUST00000174291.1
ENSMUST00000167888.2 ENSMUST00000172662.1 ENSMUST00000173270.1 ENSMUST00000174083.1 |
Stard10
|
START domain containing 10 |
| chr8_+_87472805 | 1.12 |
ENSMUST00000180700.2
ENSMUST00000182174.1 ENSMUST00000181159.1 |
Gm2694
|
predicted gene 2694 |
| chr14_-_33447142 | 1.11 |
ENSMUST00000111944.3
ENSMUST00000022504.5 ENSMUST00000111945.2 |
Mapk8
|
mitogen-activated protein kinase 8 |
| chr2_+_122234749 | 1.11 |
ENSMUST00000110551.3
|
Sord
|
sorbitol dehydrogenase |
| chr7_-_90457167 | 1.08 |
ENSMUST00000032844.5
|
Tmem126a
|
transmembrane protein 126A |
| chr7_-_31110997 | 1.08 |
ENSMUST00000039435.8
|
Hpn
|
hepsin |
| chr14_-_55784007 | 1.05 |
ENSMUST00000002398.7
|
Adcy4
|
adenylate cyclase 4 |
| chr12_-_103242143 | 1.04 |
ENSMUST00000074416.3
|
Prima1
|
proline rich membrane anchor 1 |
| chr14_-_55784027 | 1.03 |
ENSMUST00000170223.1
|
Adcy4
|
adenylate cyclase 4 |
| chr8_+_119394866 | 1.02 |
ENSMUST00000098367.4
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr7_-_31111148 | 1.01 |
ENSMUST00000164929.1
|
Hpn
|
hepsin |
| chr15_+_52040107 | 1.00 |
ENSMUST00000090025.4
|
Aard
|
alanine and arginine rich domain containing protein |
| chr2_-_168742100 | 1.00 |
ENSMUST00000109177.1
|
Atp9a
|
ATPase, class II, type 9A |
| chr11_-_77489666 | 0.99 |
ENSMUST00000037593.7
ENSMUST00000092892.3 |
Ankrd13b
|
ankyrin repeat domain 13b |
| chr14_+_45219993 | 0.97 |
ENSMUST00000146150.1
|
Gpr137c
|
G protein-coupled receptor 137C |
| chr15_-_76009440 | 0.96 |
ENSMUST00000170153.1
|
Fam83h
|
family with sequence similarity 83, member H |
| chr5_+_92897981 | 0.95 |
ENSMUST00000113051.2
|
Shroom3
|
shroom family member 3 |
| chr5_-_121385599 | 0.94 |
ENSMUST00000146185.1
ENSMUST00000042312.7 |
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr6_+_54816906 | 0.94 |
ENSMUST00000079869.6
|
Znrf2
|
zinc and ring finger 2 |
| chr4_-_129239165 | 0.93 |
ENSMUST00000097873.3
|
C77080
|
expressed sequence C77080 |
| chr1_+_172482199 | 0.92 |
ENSMUST00000135267.1
ENSMUST00000052629.6 ENSMUST00000111235.2 |
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr1_+_21218575 | 0.89 |
ENSMUST00000027065.5
ENSMUST00000027064.7 |
Tmem14a
|
transmembrane protein 14A |
| chr19_+_6418731 | 0.89 |
ENSMUST00000113462.1
ENSMUST00000077182.6 ENSMUST00000113461.1 |
Nrxn2
|
neurexin II |
| chr4_-_89294608 | 0.89 |
ENSMUST00000107131.1
|
Cdkn2a
|
cyclin-dependent kinase inhibitor 2A |
| chr11_-_23665862 | 0.89 |
ENSMUST00000020523.3
|
Pex13
|
peroxisomal biogenesis factor 13 |
| chr1_+_164249233 | 0.88 |
ENSMUST00000169394.1
|
Slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr1_+_23762003 | 0.88 |
ENSMUST00000140583.1
|
B3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr11_+_120721452 | 0.87 |
ENSMUST00000018156.5
|
Rac3
|
RAS-related C3 botulinum substrate 3 |
| chr8_+_87472838 | 0.87 |
ENSMUST00000180806.2
|
Gm2694
|
predicted gene 2694 |
| chr4_-_117133953 | 0.87 |
ENSMUST00000076859.5
|
Plk3
|
polo-like kinase 3 |
| chr1_+_60409612 | 0.87 |
ENSMUST00000052332.8
|
Abi2
|
abl-interactor 2 |
| chr4_+_155993143 | 0.85 |
ENSMUST00000097734.4
|
Sdf4
|
stromal cell derived factor 4 |
| chr6_-_88518760 | 0.83 |
ENSMUST00000032168.5
|
Sec61a1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr3_-_95882232 | 0.82 |
ENSMUST00000161866.1
|
Gm129
|
predicted gene 129 |
| chr9_+_103112072 | 0.82 |
ENSMUST00000035155.6
|
Rab6b
|
RAB6B, member RAS oncogene family |
| chr7_-_27166413 | 0.81 |
ENSMUST00000108382.1
|
Egln2
|
EGL nine homolog 2 (C. elegans) |
| chr3_-_88177671 | 0.79 |
ENSMUST00000181837.1
|
1700113A16Rik
|
RIKEN cDNA 1700113A16 gene |
| chr4_-_11966368 | 0.78 |
ENSMUST00000056050.4
ENSMUST00000108299.1 ENSMUST00000108297.2 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr7_+_35119285 | 0.77 |
ENSMUST00000042985.9
|
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chrX_+_140664908 | 0.74 |
ENSMUST00000112990.1
ENSMUST00000112988.1 |
Mid2
|
midline 2 |
| chr1_+_172481788 | 0.74 |
ENSMUST00000127052.1
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr11_+_120348678 | 0.73 |
ENSMUST00000143813.1
|
0610009L18Rik
|
RIKEN cDNA 0610009L18 gene |
| chr10_+_80141457 | 0.72 |
ENSMUST00000105367.1
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr11_+_103966716 | 0.69 |
ENSMUST00000057921.3
ENSMUST00000063347.5 |
Arf2
|
ADP-ribosylation factor 2 |
| chr18_-_68300194 | 0.68 |
ENSMUST00000152193.1
|
Fam210a
|
family with sequence similarity 210, member A |
| chr9_-_114933929 | 0.68 |
ENSMUST00000146623.1
|
Gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr2_-_164190601 | 0.67 |
ENSMUST00000051272.7
|
Wfdc12
|
WAP four-disulfide core domain 12 |
| chr9_-_53610329 | 0.66 |
ENSMUST00000034547.5
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr1_-_60566708 | 0.65 |
ENSMUST00000027168.5
ENSMUST00000090293.4 ENSMUST00000140485.1 |
Raph1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr11_+_9048575 | 0.65 |
ENSMUST00000043285.4
|
Gm11992
|
predicted gene 11992 |
| chr10_+_79854618 | 0.64 |
ENSMUST00000165704.1
|
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr8_+_71568866 | 0.64 |
ENSMUST00000034267.4
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr1_-_30949756 | 0.64 |
ENSMUST00000076587.3
ENSMUST00000027232.7 |
Ptp4a1
|
protein tyrosine phosphatase 4a1 |
| chr1_+_120340569 | 0.63 |
ENSMUST00000037286.8
|
C1ql2
|
complement component 1, q subcomponent-like 2 |
| chr18_-_68300329 | 0.63 |
ENSMUST00000042852.6
|
Fam210a
|
family with sequence similarity 210, member A |
| chr11_+_120721543 | 0.63 |
ENSMUST00000142229.1
|
Rac3
|
RAS-related C3 botulinum substrate 3 |
| chr11_+_60105079 | 0.62 |
ENSMUST00000132012.1
|
Rai1
|
retinoic acid induced 1 |
| chr5_+_100846364 | 0.62 |
ENSMUST00000092990.3
ENSMUST00000145612.1 |
Agpat9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr16_+_92058270 | 0.62 |
ENSMUST00000047429.8
ENSMUST00000113975.2 |
Mrps6
Slc5a3
|
mitochondrial ribosomal protein S6 solute carrier family 5 (inositol transporters), member 3 |
| chr12_-_75735729 | 0.61 |
ENSMUST00000021450.4
|
Sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr15_-_91573181 | 0.59 |
ENSMUST00000109283.1
|
Slc2a13
|
solute carrier family 2 (facilitated glucose transporter), member 13 |
| chr8_-_93810225 | 0.56 |
ENSMUST00000181864.1
|
Gm26843
|
predicted gene, 26843 |
| chr10_+_80142295 | 0.55 |
ENSMUST00000003156.8
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr14_-_121797670 | 0.53 |
ENSMUST00000100299.3
|
Dock9
|
dedicator of cytokinesis 9 |
| chr7_-_34655500 | 0.53 |
ENSMUST00000032709.1
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr1_+_164249052 | 0.53 |
ENSMUST00000159230.1
|
Slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr10_+_79854658 | 0.52 |
ENSMUST00000171599.1
ENSMUST00000095457.4 |
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr4_+_129513581 | 0.52 |
ENSMUST00000062356.6
|
Marcksl1
|
MARCKS-like 1 |
| chr1_+_23761749 | 0.52 |
ENSMUST00000144602.1
|
B3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr4_+_126753770 | 0.52 |
ENSMUST00000102607.3
ENSMUST00000047431.4 ENSMUST00000132660.1 |
AU040320
|
expressed sequence AU040320 |
| chr11_-_115419917 | 0.51 |
ENSMUST00000106537.1
ENSMUST00000043931.2 ENSMUST00000073791.3 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d |
| chr10_+_69212676 | 0.51 |
ENSMUST00000167384.1
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr5_-_24842579 | 0.50 |
ENSMUST00000030787.8
|
Rheb
|
Ras homolog enriched in brain |
| chr12_+_80518990 | 0.49 |
ENSMUST00000021558.6
|
Galnt16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr5_-_122502192 | 0.48 |
ENSMUST00000179939.1
ENSMUST00000177974.1 ENSMUST00000031423.8 |
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr2_+_3284202 | 0.48 |
ENSMUST00000081932.6
|
Nmt2
|
N-myristoyltransferase 2 |
| chr11_-_100939357 | 0.47 |
ENSMUST00000092671.5
ENSMUST00000103114.1 |
Stat3
|
signal transducer and activator of transcription 3 |
| chr7_-_30729505 | 0.46 |
ENSMUST00000006478.8
|
Tmem147
|
transmembrane protein 147 |
| chr11_-_93955718 | 0.46 |
ENSMUST00000072566.4
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr9_-_114933811 | 0.46 |
ENSMUST00000084853.3
|
Gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr4_+_129058133 | 0.46 |
ENSMUST00000030584.4
ENSMUST00000168461.1 ENSMUST00000152565.1 |
Rnf19b
|
ring finger protein 19B |
| chr1_+_164249023 | 0.45 |
ENSMUST00000044021.5
|
Slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr4_-_126753372 | 0.45 |
ENSMUST00000030637.7
ENSMUST00000106116.1 |
Ncdn
|
neurochondrin |
| chr10_+_80142358 | 0.44 |
ENSMUST00000105366.1
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr14_-_31128924 | 0.44 |
ENSMUST00000064032.4
ENSMUST00000049732.5 ENSMUST00000090205.3 |
Smim4
|
small itegral membrane protein 4 |
| chr10_-_112928974 | 0.44 |
ENSMUST00000099276.2
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr1_+_88406956 | 0.44 |
ENSMUST00000027518.5
|
Spp2
|
secreted phosphoprotein 2 |
| chr6_+_22288221 | 0.43 |
ENSMUST00000128245.1
ENSMUST00000031681.3 ENSMUST00000148639.1 |
Wnt16
|
wingless-related MMTV integration site 16 |
| chr1_+_119526125 | 0.42 |
ENSMUST00000183952.1
|
TMEM185B
|
Transmembrane protein 185B |
| chr6_-_88875035 | 0.42 |
ENSMUST00000145944.1
|
Podxl2
|
podocalyxin-like 2 |
| chr4_+_101419696 | 0.41 |
ENSMUST00000131397.1
ENSMUST00000133055.1 |
Ak4
|
adenylate kinase 4 |
| chr3_+_90052814 | 0.41 |
ENSMUST00000160640.1
ENSMUST00000029552.6 ENSMUST00000162114.1 ENSMUST00000068798.6 |
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr5_-_139484420 | 0.41 |
ENSMUST00000150992.1
|
Zfand2a
|
zinc finger, AN1-type domain 2A |
| chr14_-_67933512 | 0.41 |
ENSMUST00000039135.4
|
Dock5
|
dedicator of cytokinesis 5 |
| chr10_+_127739516 | 0.40 |
ENSMUST00000054287.7
|
Zbtb39
|
zinc finger and BTB domain containing 39 |
| chr1_+_182763961 | 0.40 |
ENSMUST00000153348.1
|
Susd4
|
sushi domain containing 4 |
| chr1_-_52500679 | 0.40 |
ENSMUST00000069792.7
|
Nab1
|
Ngfi-A binding protein 1 |
| chr3_+_40708855 | 0.39 |
ENSMUST00000091184.6
|
Slc25a31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 |
| chr8_-_94012558 | 0.39 |
ENSMUST00000053766.6
|
Amfr
|
autocrine motility factor receptor |
| chr16_+_33829624 | 0.39 |
ENSMUST00000115028.3
|
Itgb5
|
integrin beta 5 |
| chr9_+_60794468 | 0.39 |
ENSMUST00000050183.6
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr4_+_138304723 | 0.39 |
ENSMUST00000030538.4
|
Ddost
|
dolichyl-di-phosphooligosaccharide-protein glycotransferase |
| chr8_+_119612747 | 0.38 |
ENSMUST00000098361.2
|
Adad2
|
adenosine deaminase domain containing 2 |
| chr3_-_95882031 | 0.37 |
ENSMUST00000161994.1
|
Gm129
|
predicted gene 129 |
| chr2_+_74762980 | 0.37 |
ENSMUST00000047793.4
|
Hoxd1
|
homeobox D1 |
| chr10_-_61452658 | 0.37 |
ENSMUST00000167087.1
ENSMUST00000020288.7 |
Eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr1_+_36511867 | 0.37 |
ENSMUST00000001166.7
ENSMUST00000097776.3 |
Cnnm3
|
cyclin M3 |
| chr3_-_95882193 | 0.37 |
ENSMUST00000159863.1
ENSMUST00000159739.1 ENSMUST00000036418.3 |
Gm129
|
predicted gene 129 |
| chr17_-_27728889 | 0.36 |
ENSMUST00000167489.1
ENSMUST00000138970.1 ENSMUST00000114870.1 ENSMUST00000025054.2 |
Spdef
|
SAM pointed domain containing ets transcription factor |
| chr2_+_128967383 | 0.36 |
ENSMUST00000110320.2
ENSMUST00000110319.2 |
Zc3h6
|
zinc finger CCCH type containing 6 |
| chr6_-_88874597 | 0.35 |
ENSMUST00000061262.4
ENSMUST00000140455.1 ENSMUST00000145780.1 |
Podxl2
|
podocalyxin-like 2 |
| chr1_-_16519284 | 0.35 |
ENSMUST00000162751.1
ENSMUST00000027052.6 ENSMUST00000149320.2 |
Stau2
|
staufen (RNA binding protein) homolog 2 (Drosophila) |
| chr9_-_56418023 | 0.35 |
ENSMUST00000061552.8
|
C230081A13Rik
|
RIKEN cDNA C230081A13 gene |
| chr8_+_87473116 | 0.35 |
ENSMUST00000182650.1
ENSMUST00000182758.1 ENSMUST00000181898.1 |
Gm2694
|
predicted gene 2694 |
| chr10_+_69212634 | 0.34 |
ENSMUST00000020101.5
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr3_+_89418443 | 0.34 |
ENSMUST00000039110.5
ENSMUST00000125036.1 ENSMUST00000154791.1 ENSMUST00000128238.1 ENSMUST00000107417.2 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr10_+_128909866 | 0.33 |
ENSMUST00000026407.7
|
Cd63
|
CD63 antigen |
| chr17_-_46031813 | 0.32 |
ENSMUST00000024747.7
|
Vegfa
|
vascular endothelial growth factor A |
| chr1_-_152766281 | 0.32 |
ENSMUST00000111859.1
ENSMUST00000148865.1 |
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr7_+_80261202 | 0.31 |
ENSMUST00000117989.1
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr2_-_93334467 | 0.31 |
ENSMUST00000111265.2
|
Tspan18
|
tetraspanin 18 |
| chr11_+_115420059 | 0.30 |
ENSMUST00000103035.3
|
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr18_+_62324204 | 0.30 |
ENSMUST00000027560.6
|
Htr4
|
5 hydroxytryptamine (serotonin) receptor 4 |
| chr9_+_75410145 | 0.30 |
ENSMUST00000180533.1
ENSMUST00000180574.1 |
4933433G15Rik
|
RIKEN cDNA 4933433G15 gene |
| chr5_+_67260794 | 0.30 |
ENSMUST00000161369.1
|
Tmem33
|
transmembrane protein 33 |
| chr5_+_135168382 | 0.29 |
ENSMUST00000111187.3
ENSMUST00000111188.1 |
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr8_-_47289394 | 0.29 |
ENSMUST00000079195.5
|
Stox2
|
storkhead box 2 |
| chr2_+_24949747 | 0.28 |
ENSMUST00000028350.3
|
Zmynd19
|
zinc finger, MYND domain containing 19 |
| chr9_+_21165714 | 0.28 |
ENSMUST00000039413.8
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr12_+_87200524 | 0.25 |
ENSMUST00000182869.1
|
Samd15
|
sterile alpha motif domain containing 15 |
| chr1_-_16519201 | 0.25 |
ENSMUST00000159558.1
ENSMUST00000054668.6 ENSMUST00000162627.1 ENSMUST00000162007.1 ENSMUST00000128957.2 ENSMUST00000115359.3 ENSMUST00000151888.1 |
Stau2
|
staufen (RNA binding protein) homolog 2 (Drosophila) |
| chr6_-_28421680 | 0.24 |
ENSMUST00000090511.3
|
Gcc1
|
golgi coiled coil 1 |
| chr19_+_22139028 | 0.24 |
ENSMUST00000099569.2
ENSMUST00000087576.4 ENSMUST00000074770.5 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr6_-_11907419 | 0.24 |
ENSMUST00000031637.5
|
Ndufa4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 |
| chr11_-_106388066 | 0.24 |
ENSMUST00000106813.2
ENSMUST00000141146.1 |
Icam2
|
intercellular adhesion molecule 2 |
| chr9_+_109096659 | 0.23 |
ENSMUST00000130366.1
|
Plxnb1
|
plexin B1 |
| chr7_+_29519158 | 0.22 |
ENSMUST00000141713.1
|
4932431P20Rik
|
RIKEN cDNA 4932431P20 gene |
| chr1_-_93445642 | 0.22 |
ENSMUST00000042498.7
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr6_+_17306335 | 0.20 |
ENSMUST00000007799.6
ENSMUST00000115456.1 ENSMUST00000115455.2 ENSMUST00000130505.1 |
Cav1
|
caveolin 1, caveolae protein |
| chr12_+_4133394 | 0.20 |
ENSMUST00000152065.1
ENSMUST00000127756.1 |
Adcy3
|
adenylate cyclase 3 |
| chr8_+_4349588 | 0.19 |
ENSMUST00000110982.1
ENSMUST00000024004.7 |
Ccl25
|
chemokine (C-C motif) ligand 25 |
| chr10_+_128083273 | 0.19 |
ENSMUST00000026459.5
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr15_+_81744848 | 0.18 |
ENSMUST00000109554.1
|
Zc3h7b
|
zinc finger CCCH type containing 7B |
| chr1_+_92906959 | 0.18 |
ENSMUST00000060913.6
|
Dusp28
|
dual specificity phosphatase 28 |
| chr16_+_94085226 | 0.17 |
ENSMUST00000072182.7
|
Sim2
|
single-minded homolog 2 (Drosophila) |
| chr11_+_115420138 | 0.17 |
ENSMUST00000106533.1
ENSMUST00000123345.1 |
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr1_-_152766323 | 0.16 |
ENSMUST00000111857.1
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr2_+_74711911 | 0.16 |
ENSMUST00000111983.2
|
Hoxd3
|
homeobox D3 |
| chr2_+_167503089 | 0.15 |
ENSMUST00000078050.6
|
Rnf114
|
ring finger protein 114 |
| chr4_+_118428078 | 0.15 |
ENSMUST00000006557.6
ENSMUST00000167636.1 ENSMUST00000102673.4 |
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr7_-_44892358 | 0.14 |
ENSMUST00000003049.6
|
Med25
|
mediator of RNA polymerase II transcription, subunit 25 homolog (yeast) |
| chr7_+_79500081 | 0.14 |
ENSMUST00000181511.2
ENSMUST00000182937.1 |
AI854517
|
expressed sequence AI854517 |
| chr15_-_39112642 | 0.14 |
ENSMUST00000022908.8
|
Slc25a32
|
solute carrier family 25, member 32 |
| chr1_-_187143072 | 0.14 |
ENSMUST00000184543.1
|
Spata17
|
spermatogenesis associated 17 |
| chr4_+_85205120 | 0.14 |
ENSMUST00000107188.3
|
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr1_+_178187721 | 0.14 |
ENSMUST00000159284.1
|
Desi2
|
desumoylating isopeptidase 2 |
| chr11_+_74830920 | 0.14 |
ENSMUST00000000291.2
|
Mnt
|
max binding protein |
| chr9_+_22156838 | 0.13 |
ENSMUST00000123680.1
|
Pigyl
|
phosphatidylinositol glycan anchor biosynthesis, class Y-like |
| chr8_+_4248188 | 0.13 |
ENSMUST00000110993.1
|
Gm14378
|
predicted gene 14378 |
| chr14_-_31830402 | 0.13 |
ENSMUST00000014640.7
|
Ankrd28
|
ankyrin repeat domain 28 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hey2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.6 | 1.8 | GO:1904020 | positive regulation of adenosine receptor signaling pathway(GO:0060168) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.6 | 1.7 | GO:0018894 | coumarin metabolic process(GO:0009804) dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.5 | 2.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.5 | 3.8 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.5 | 1.4 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.4 | 1.1 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.4 | 1.9 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.4 | 1.1 | GO:0046370 | hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) fructose biosynthetic process(GO:0046370) |
| 0.3 | 1.0 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.3 | 0.6 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.3 | 7.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 0.8 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.3 | 1.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.3 | 2.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 3.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 1.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.2 | 3.9 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 6.0 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 1.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 1.5 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.2 | 0.9 | GO:2000434 | positive regulation of macrophage apoptotic process(GO:2000111) regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.2 | 0.5 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) |
| 0.2 | 0.5 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.2 | 3.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 0.6 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.9 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.8 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.4 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.1 | 0.5 | GO:1902724 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 1.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.9 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.3 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.1 | 0.8 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 0.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.6 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 1.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.9 | GO:0090166 | Golgi disassembly(GO:0090166) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 1.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 0.4 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 2.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.1 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.1 | 0.3 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 1.0 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.4 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.8 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 | 3.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 2.0 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.1 | 0.3 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.1 | 0.3 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 1.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.6 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.8 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 1.9 | GO:0030811 | regulation of glycolytic process(GO:0006110) regulation of nucleotide catabolic process(GO:0030811) |
| 0.0 | 0.4 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 2.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 3.8 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.1 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 1.0 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.4 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.5 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 1.3 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 1.0 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 1.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.9 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.1 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0038003 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:0006544 | glycine metabolic process(GO:0006544) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.6 | 3.5 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 0.8 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.2 | 1.9 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 1.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.4 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.1 | 0.9 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.5 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 4.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 1.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 1.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 8.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 2.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 3.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.7 | 3.9 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 2.2 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.5 | 1.9 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.4 | 1.8 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.3 | 3.3 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.3 | 1.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 1.9 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 1.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 1.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 0.9 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 0.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 1.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 1.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 1.7 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.2 | 0.5 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 1.7 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 2.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.7 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 2.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 1.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.9 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 2.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 3.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 2.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.8 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.5 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.1 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 1.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.8 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.0 | 1.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 2.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 5.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 4.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 8.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 4.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 3.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 3.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 6.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 3.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 2.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.7 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 3.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 2.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 4.4 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |