Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
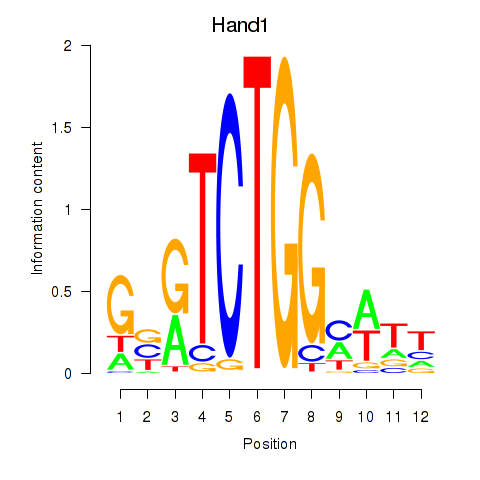
Results for Hand1
Z-value: 1.09
Transcription factors associated with Hand1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hand1
|
ENSMUSG00000037335.7 | heart and neural crest derivatives expressed 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hand1 | mm10_v2_chr11_-_57832679_57832818 | 0.68 | 4.6e-06 | Click! |
Activity profile of Hand1 motif
Sorted Z-values of Hand1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_75568648 | 6.90 |
ENSMUST00000134503.1
ENSMUST00000125770.1 ENSMUST00000128886.1 ENSMUST00000151212.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr10_+_75568630 | 6.52 |
ENSMUST00000145928.1
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr10_+_75568641 | 6.25 |
ENSMUST00000131565.1
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr7_-_17056669 | 6.15 |
ENSMUST00000037762.4
|
Hif3a
|
hypoxia inducible factor 3, alpha subunit |
| chr8_-_85365341 | 5.91 |
ENSMUST00000121972.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_-_85365317 | 5.80 |
ENSMUST00000034133.7
|
Mylk3
|
myosin light chain kinase 3 |
| chr11_+_4236411 | 5.60 |
ENSMUST00000075221.2
|
Osm
|
oncostatin M |
| chr7_+_18884679 | 4.64 |
ENSMUST00000032573.6
|
Pglyrp1
|
peptidoglycan recognition protein 1 |
| chr17_-_26199008 | 4.59 |
ENSMUST00000142410.1
ENSMUST00000120333.1 ENSMUST00000039113.7 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr2_-_121036750 | 4.48 |
ENSMUST00000023987.5
|
Epb4.2
|
erythrocyte protein band 4.2 |
| chr13_-_37049203 | 4.41 |
ENSMUST00000037491.8
|
F13a1
|
coagulation factor XIII, A1 subunit |
| chr7_+_143005638 | 4.37 |
ENSMUST00000075172.5
ENSMUST00000105923.1 |
Tspan32
|
tetraspanin 32 |
| chr2_-_121037048 | 4.25 |
ENSMUST00000102490.3
|
Epb4.2
|
erythrocyte protein band 4.2 |
| chr7_+_143005046 | 4.13 |
ENSMUST00000009396.6
|
Tspan32
|
tetraspanin 32 |
| chr6_-_41636389 | 3.91 |
ENSMUST00000031902.5
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr15_+_80097866 | 3.89 |
ENSMUST00000143928.1
|
Syngr1
|
synaptogyrin 1 |
| chr14_-_37110087 | 3.65 |
ENSMUST00000179488.1
|
2610528A11Rik
|
RIKEN cDNA 2610528A11 gene |
| chr7_-_141655319 | 3.48 |
ENSMUST00000062451.7
|
Muc6
|
mucin 6, gastric |
| chr8_+_70373541 | 3.35 |
ENSMUST00000003659.7
|
Comp
|
cartilage oligomeric matrix protein |
| chr2_-_28563362 | 3.34 |
ENSMUST00000028161.5
|
Cel
|
carboxyl ester lipase |
| chr7_+_131032061 | 3.31 |
ENSMUST00000084509.3
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr15_-_79285502 | 3.21 |
ENSMUST00000165408.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr5_-_107723954 | 3.18 |
ENSMUST00000165344.1
|
Gfi1
|
growth factor independent 1 |
| chr19_+_58728887 | 3.14 |
ENSMUST00000048644.5
|
Pnliprp1
|
pancreatic lipase related protein 1 |
| chr7_-_4752972 | 3.12 |
ENSMUST00000183971.1
ENSMUST00000182173.1 ENSMUST00000182738.1 ENSMUST00000184143.1 ENSMUST00000182111.1 ENSMUST00000182048.1 ENSMUST00000063324.7 |
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr4_-_4138817 | 2.97 |
ENSMUST00000133567.1
|
Penk
|
preproenkephalin |
| chr7_-_30445508 | 2.86 |
ENSMUST00000006828.7
|
Aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr2_+_173022360 | 2.75 |
ENSMUST00000173997.1
|
Rbm38
|
RNA binding motif protein 38 |
| chr8_+_84723003 | 2.60 |
ENSMUST00000098571.4
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene |
| chr8_+_95055094 | 2.60 |
ENSMUST00000058479.6
|
Ccdc135
|
coiled-coil domain containing 135 |
| chr12_-_8539545 | 2.52 |
ENSMUST00000095863.3
ENSMUST00000165657.1 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr17_+_48454891 | 2.47 |
ENSMUST00000074574.6
|
Unc5cl
|
unc-5 homolog C (C. elegans)-like |
| chr6_-_67535783 | 2.44 |
ENSMUST00000058178.4
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr11_-_3504766 | 2.43 |
ENSMUST00000044507.5
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr4_-_117883428 | 2.40 |
ENSMUST00000030266.5
|
B4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr6_+_30639218 | 2.40 |
ENSMUST00000031806.9
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr18_+_67343564 | 2.30 |
ENSMUST00000025404.8
|
Cidea
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector A |
| chr19_+_7268296 | 2.22 |
ENSMUST00000066646.4
|
Rcor2
|
REST corepressor 2 |
| chr6_-_126939524 | 2.22 |
ENSMUST00000144954.1
ENSMUST00000112221.1 ENSMUST00000112220.1 |
Rad51ap1
|
RAD51 associated protein 1 |
| chr7_+_16781341 | 2.19 |
ENSMUST00000108496.2
|
Slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr17_-_24658425 | 2.19 |
ENSMUST00000095544.4
|
Npw
|
neuropeptide W |
| chr1_+_75507077 | 2.14 |
ENSMUST00000037330.4
|
Inha
|
inhibin alpha |
| chr14_-_54712139 | 2.12 |
ENSMUST00000064290.6
|
Cebpe
|
CCAAT/enhancer binding protein (C/EBP), epsilon |
| chr7_-_126414855 | 2.09 |
ENSMUST00000032968.5
|
Cd19
|
CD19 antigen |
| chr19_-_42086338 | 2.06 |
ENSMUST00000051772.8
|
Morn4
|
MORN repeat containing 4 |
| chr11_-_69948145 | 2.04 |
ENSMUST00000179298.1
ENSMUST00000018710.6 ENSMUST00000135437.1 ENSMUST00000141837.2 ENSMUST00000142500.1 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr4_-_117929726 | 2.00 |
ENSMUST00000070816.2
|
Artn
|
artemin |
| chr10_+_83722865 | 1.96 |
ENSMUST00000150459.1
|
1500009L16Rik
|
RIKEN cDNA 1500009L16 gene |
| chr11_+_46235460 | 1.96 |
ENSMUST00000060185.2
|
Fndc9
|
fibronectin type III domain containing 9 |
| chr18_-_74207771 | 1.89 |
ENSMUST00000040188.8
ENSMUST00000177604.1 |
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr1_+_134182150 | 1.86 |
ENSMUST00000156873.1
|
Chi3l1
|
chitinase 3-like 1 |
| chr2_+_79255500 | 1.86 |
ENSMUST00000099972.4
|
Itga4
|
integrin alpha 4 |
| chr7_-_143094642 | 1.84 |
ENSMUST00000009390.3
|
Trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr17_+_33555719 | 1.77 |
ENSMUST00000087605.5
ENSMUST00000174695.1 |
Myo1f
|
myosin IF |
| chr10_+_127514939 | 1.77 |
ENSMUST00000035735.9
|
Ndufa4l2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr2_+_85136355 | 1.75 |
ENSMUST00000057019.7
|
Aplnr
|
apelin receptor |
| chr9_+_53771499 | 1.74 |
ENSMUST00000048670.8
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr17_+_56040350 | 1.70 |
ENSMUST00000002914.8
|
Chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr13_+_21495218 | 1.69 |
ENSMUST00000104942.1
|
AK157302
|
cDNA sequence AK157302 |
| chr1_-_193201435 | 1.67 |
ENSMUST00000043550.4
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr7_+_103937382 | 1.66 |
ENSMUST00000098189.1
|
Olfr632
|
olfactory receptor 632 |
| chr4_+_141010644 | 1.64 |
ENSMUST00000071977.8
|
Mfap2
|
microfibrillar-associated protein 2 |
| chr12_-_32208470 | 1.63 |
ENSMUST00000085469.5
|
Pik3cg
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide |
| chr14_+_54476100 | 1.63 |
ENSMUST00000164766.1
ENSMUST00000164697.1 |
Rem2
|
rad and gem related GTP binding protein 2 |
| chr15_+_79348061 | 1.57 |
ENSMUST00000163691.1
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr7_+_141061274 | 1.55 |
ENSMUST00000048002.5
|
B4galnt4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr2_+_14873656 | 1.52 |
ENSMUST00000114718.1
ENSMUST00000114719.1 |
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr4_-_42853888 | 1.50 |
ENSMUST00000107979.1
|
Gm12429
|
predicted gene 12429 |
| chr3_+_87948666 | 1.46 |
ENSMUST00000005019.5
|
Crabp2
|
cellular retinoic acid binding protein II |
| chr4_+_119637704 | 1.45 |
ENSMUST00000024015.2
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr1_-_176807124 | 1.44 |
ENSMUST00000057037.7
|
Cep170
|
centrosomal protein 170 |
| chr11_-_102925086 | 1.44 |
ENSMUST00000021311.9
|
Kif18b
|
kinesin family member 18B |
| chr7_+_43440782 | 1.44 |
ENSMUST00000040227.1
|
Cldnd2
|
claudin domain containing 2 |
| chr7_+_81858993 | 1.41 |
ENSMUST00000041890.1
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr8_+_57511833 | 1.40 |
ENSMUST00000067925.6
|
Hmgb2
|
high mobility group box 2 |
| chr3_-_90389884 | 1.39 |
ENSMUST00000029541.5
|
Slc27a3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr4_+_135120640 | 1.36 |
ENSMUST00000056977.7
|
Runx3
|
runt related transcription factor 3 |
| chr15_+_81235499 | 1.36 |
ENSMUST00000166855.1
|
Mchr1
|
melanin-concentrating hormone receptor 1 |
| chr19_-_47692042 | 1.36 |
ENSMUST00000026045.7
ENSMUST00000086923.5 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr4_-_45084538 | 1.35 |
ENSMUST00000052236.6
|
Fbxo10
|
F-box protein 10 |
| chr9_-_44454757 | 1.35 |
ENSMUST00000047740.2
|
Upk2
|
uroplakin 2 |
| chr12_-_32208609 | 1.35 |
ENSMUST00000053215.7
|
Pik3cg
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide |
| chr6_+_127887582 | 1.34 |
ENSMUST00000032501.4
|
Tspan11
|
tetraspanin 11 |
| chr18_+_60963517 | 1.32 |
ENSMUST00000115295.2
ENSMUST00000039904.6 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr7_+_30421724 | 1.29 |
ENSMUST00000108176.1
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr7_+_28788955 | 1.28 |
ENSMUST00000059857.7
|
Rinl
|
Ras and Rab interactor-like |
| chr5_+_47984793 | 1.28 |
ENSMUST00000170109.2
ENSMUST00000174421.1 ENSMUST00000173702.1 ENSMUST00000173107.1 |
Slit2
|
slit homolog 2 (Drosophila) |
| chr7_+_43797567 | 1.27 |
ENSMUST00000085461.2
|
Klk8
|
kallikrein related-peptidase 8 |
| chr11_-_114934351 | 1.26 |
ENSMUST00000106581.1
ENSMUST00000074300.2 |
Cd300lb
|
CD300 antigen like family member B |
| chr5_+_108132885 | 1.24 |
ENSMUST00000047677.7
|
Ccdc18
|
coiled-coil domain containing 18 |
| chr5_-_113830422 | 1.24 |
ENSMUST00000100874.4
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr11_+_3330781 | 1.22 |
ENSMUST00000136536.1
ENSMUST00000093399.4 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr7_+_24862193 | 1.21 |
ENSMUST00000052897.4
ENSMUST00000170837.2 |
Gm9844
Gm9844
|
predicted pseudogene 9844 predicted pseudogene 9844 |
| chr7_+_45215753 | 1.20 |
ENSMUST00000033060.6
ENSMUST00000155313.1 ENSMUST00000107801.1 |
Tead2
|
TEA domain family member 2 |
| chr4_+_148643317 | 1.19 |
ENSMUST00000105698.2
|
Gm572
|
predicted gene 572 |
| chr17_+_47596061 | 1.17 |
ENSMUST00000182539.1
|
Ccnd3
|
cyclin D3 |
| chr14_+_32321987 | 1.16 |
ENSMUST00000022480.7
|
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr4_+_115088708 | 1.15 |
ENSMUST00000171877.1
ENSMUST00000177647.1 ENSMUST00000106548.2 ENSMUST00000030488.2 |
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr17_-_26095487 | 1.14 |
ENSMUST00000025007.5
|
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr12_-_76822510 | 1.13 |
ENSMUST00000021459.7
|
Rab15
|
RAB15, member RAS oncogene family |
| chr8_+_93810832 | 1.12 |
ENSMUST00000034198.8
ENSMUST00000125716.1 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr1_+_135783065 | 1.12 |
ENSMUST00000132795.1
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr3_+_108383829 | 1.12 |
ENSMUST00000090561.3
ENSMUST00000102629.1 ENSMUST00000128089.1 |
Psrc1
|
proline/serine-rich coiled-coil 1 |
| chr5_-_138172383 | 1.11 |
ENSMUST00000000505.9
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr1_-_65123108 | 1.10 |
ENSMUST00000050047.3
ENSMUST00000148020.1 |
D630023F18Rik
|
RIKEN cDNA D630023F18 gene |
| chr7_-_119479249 | 1.10 |
ENSMUST00000033263.4
|
Umod
|
uromodulin |
| chr7_+_28693032 | 1.07 |
ENSMUST00000151227.1
ENSMUST00000108281.1 |
Fbxo27
|
F-box protein 27 |
| chr8_-_88636117 | 1.06 |
ENSMUST00000034087.7
|
Snx20
|
sorting nexin 20 |
| chr4_-_133872997 | 1.06 |
ENSMUST00000137486.2
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr18_-_52529692 | 1.06 |
ENSMUST00000025409.7
|
Lox
|
lysyl oxidase |
| chr12_+_113156403 | 1.06 |
ENSMUST00000049271.8
|
4930427A07Rik
|
RIKEN cDNA 4930427A07 gene |
| chr1_+_86021935 | 1.05 |
ENSMUST00000052854.6
ENSMUST00000125083.1 ENSMUST00000152501.1 ENSMUST00000113344.1 ENSMUST00000130504.1 ENSMUST00000153247.2 |
Spata3
|
spermatogenesis associated 3 |
| chr11_-_121229095 | 1.05 |
ENSMUST00000137299.1
ENSMUST00000169393.1 |
BC017643
|
cDNA sequence BC017643 |
| chr2_+_149830840 | 1.05 |
ENSMUST00000109934.1
ENSMUST00000140870.1 |
Syndig1
|
synapse differentiation inducing 1 |
| chr11_+_95337012 | 1.04 |
ENSMUST00000037502.6
|
Fam117a
|
family with sequence similarity 117, member A |
| chr6_+_78380700 | 1.04 |
ENSMUST00000101272.1
|
Reg3a
|
regenerating islet-derived 3 alpha |
| chr1_-_120270253 | 1.04 |
ENSMUST00000112639.1
|
Steap3
|
STEAP family member 3 |
| chr2_-_118728430 | 1.00 |
ENSMUST00000102524.1
|
Plcb2
|
phospholipase C, beta 2 |
| chr7_-_98112618 | 0.98 |
ENSMUST00000153657.1
|
Myo7a
|
myosin VIIA |
| chrX_-_7188713 | 0.98 |
ENSMUST00000004428.7
|
Clcn5
|
chloride channel 5 |
| chr11_+_69914179 | 0.97 |
ENSMUST00000057884.5
|
Gps2
|
G protein pathway suppressor 2 |
| chr13_-_117025505 | 0.97 |
ENSMUST00000022239.6
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr18_-_52529847 | 0.95 |
ENSMUST00000171470.1
|
Lox
|
lysyl oxidase |
| chr17_-_55915870 | 0.95 |
ENSMUST00000074828.4
|
Rpl21-ps6
|
ribosomal protein L21, pseudogene 6 |
| chr15_-_81960851 | 0.95 |
ENSMUST00000071462.6
ENSMUST00000023112.5 |
Pmm1
|
phosphomannomutase 1 |
| chr11_-_8950539 | 0.94 |
ENSMUST00000178195.1
|
Pkd1l1
|
polycystic kidney disease 1 like 1 |
| chr7_-_79131018 | 0.93 |
ENSMUST00000032827.7
|
Hapln3
|
hyaluronan and proteoglycan link protein 3 |
| chr16_-_88751628 | 0.92 |
ENSMUST00000053149.2
|
Krtap13
|
keratin associated protein 13 |
| chr7_-_127993831 | 0.91 |
ENSMUST00000033056.3
|
Pycard
|
PYD and CARD domain containing |
| chr7_-_134232005 | 0.90 |
ENSMUST00000134504.1
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr3_+_96161981 | 0.90 |
ENSMUST00000054356.9
|
Mtmr11
|
myotubularin related protein 11 |
| chr5_+_34999046 | 0.89 |
ENSMUST00000114281.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr11_-_121229293 | 0.89 |
ENSMUST00000106115.1
ENSMUST00000038709.7 ENSMUST00000147490.1 |
BC017643
|
cDNA sequence BC017643 |
| chr7_-_4725082 | 0.88 |
ENSMUST00000086363.4
ENSMUST00000086364.4 |
Tmem150b
|
transmembrane protein 150B |
| chr2_+_131127280 | 0.88 |
ENSMUST00000099349.3
ENSMUST00000100763.2 |
Hspa12b
|
heat shock protein 12B |
| chr3_+_90341654 | 0.88 |
ENSMUST00000049382.4
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr9_+_69454066 | 0.87 |
ENSMUST00000134907.1
|
Anxa2
|
annexin A2 |
| chr16_+_92478743 | 0.87 |
ENSMUST00000160494.1
|
2410124H12Rik
|
RIKEN cDNA 2410124H12 gene |
| chr5_-_77310049 | 0.87 |
ENSMUST00000047860.8
|
Noa1
|
nitric oxide associated 1 |
| chr5_-_65091584 | 0.86 |
ENSMUST00000043352.4
|
Tmem156
|
transmembrane protein 156 |
| chr12_-_27342696 | 0.84 |
ENSMUST00000079063.5
|
Sox11
|
SRY-box containing gene 11 |
| chr15_-_75905349 | 0.84 |
ENSMUST00000127550.1
|
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chrX_+_164139321 | 0.84 |
ENSMUST00000112271.3
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr16_-_58718724 | 0.83 |
ENSMUST00000089318.3
|
Gpr15
|
G protein-coupled receptor 15 |
| chr14_-_40893222 | 0.82 |
ENSMUST00000096000.3
|
Sh2d4b
|
SH2 domain containing 4B |
| chr14_+_33954020 | 0.82 |
ENSMUST00000035695.8
|
Rbp3
|
retinol binding protein 3, interstitial |
| chr6_-_87496279 | 0.81 |
ENSMUST00000101197.2
|
Arhgap25
|
Rho GTPase activating protein 25 |
| chr10_-_62880014 | 0.81 |
ENSMUST00000050826.7
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr15_-_98607611 | 0.81 |
ENSMUST00000096224.4
|
Adcy6
|
adenylate cyclase 6 |
| chr5_+_19907502 | 0.80 |
ENSMUST00000101558.3
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr3_-_153725062 | 0.80 |
ENSMUST00000064460.5
|
St6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr5_-_124354671 | 0.80 |
ENSMUST00000031341.4
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr15_+_85510812 | 0.80 |
ENSMUST00000079690.2
|
Gm4825
|
predicted pseudogene 4825 |
| chr11_+_31832660 | 0.79 |
ENSMUST00000132857.1
|
Gm12107
|
predicted gene 12107 |
| chr4_+_134315112 | 0.78 |
ENSMUST00000105875.1
ENSMUST00000030638.6 |
Trim63
|
tripartite motif-containing 63 |
| chr15_+_77477044 | 0.78 |
ENSMUST00000060551.2
ENSMUST00000119997.1 |
Apol10a
|
apolipoprotein L 10A |
| chr15_+_58510037 | 0.77 |
ENSMUST00000161028.1
|
Fer1l6
|
fer-1-like 6 (C. elegans) |
| chr16_-_3718105 | 0.77 |
ENSMUST00000023180.7
ENSMUST00000100222.2 |
Mefv
|
Mediterranean fever |
| chr2_-_126618655 | 0.77 |
ENSMUST00000028838.4
|
Hdc
|
histidine decarboxylase |
| chr1_+_45795485 | 0.76 |
ENSMUST00000147308.1
|
Wdr75
|
WD repeat domain 75 |
| chr7_-_45366714 | 0.76 |
ENSMUST00000107779.1
|
Ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr5_-_138171813 | 0.75 |
ENSMUST00000155902.1
ENSMUST00000148879.1 |
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr13_+_21180179 | 0.75 |
ENSMUST00000021761.5
|
Trim27
|
tripartite motif-containing 27 |
| chr11_+_69125896 | 0.75 |
ENSMUST00000021268.2
|
Aloxe3
|
arachidonate lipoxygenase 3 |
| chr9_+_69453620 | 0.75 |
ENSMUST00000034756.8
ENSMUST00000123470.1 |
Anxa2
|
annexin A2 |
| chr4_+_130913264 | 0.75 |
ENSMUST00000156225.1
ENSMUST00000156742.1 |
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr1_+_135729147 | 0.75 |
ENSMUST00000027677.7
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr19_+_23723279 | 0.74 |
ENSMUST00000067077.1
|
Gm9938
|
predicted gene 9938 |
| chr7_-_118995211 | 0.73 |
ENSMUST00000008878.8
|
Gprc5b
|
G protein-coupled receptor, family C, group 5, member B |
| chr3_+_109123104 | 0.73 |
ENSMUST00000029477.6
|
Slc25a24
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 24 |
| chr7_+_30423266 | 0.73 |
ENSMUST00000046177.8
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr9_+_107547288 | 0.72 |
ENSMUST00000010188.7
|
Zmynd10
|
zinc finger, MYND domain containing 10 |
| chr2_+_118663235 | 0.72 |
ENSMUST00000099557.3
|
Pak6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr4_-_42034726 | 0.71 |
ENSMUST00000084677.2
|
Gm21093
|
predicted gene, 21093 |
| chr10_-_61784014 | 0.71 |
ENSMUST00000020283.4
|
H2afy2
|
H2A histone family, member Y2 |
| chr16_-_19515634 | 0.71 |
ENSMUST00000054606.1
|
Olfr167
|
olfactory receptor 167 |
| chr7_-_19604444 | 0.70 |
ENSMUST00000086041.5
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chr2_+_154551771 | 0.70 |
ENSMUST00000104928.1
|
Actl10
|
actin-like 10 |
| chr16_-_36784924 | 0.70 |
ENSMUST00000168279.1
ENSMUST00000164579.1 ENSMUST00000023616.2 |
Slc15a2
|
solute carrier family 15 (H+/peptide transporter), member 2 |
| chr13_+_3478226 | 0.70 |
ENSMUST00000181708.1
ENSMUST00000180836.1 ENSMUST00000180567.1 |
2810429I04Rik
|
RIKEN cDNA 2810429I04 gene |
| chr15_+_81936753 | 0.69 |
ENSMUST00000038757.7
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr17_+_46650328 | 0.69 |
ENSMUST00000043464.7
|
Cul7
|
cullin 7 |
| chr15_-_63997969 | 0.68 |
ENSMUST00000164532.1
|
Fam49b
|
family with sequence similarity 49, member B |
| chr6_-_128355826 | 0.68 |
ENSMUST00000001562.6
|
Tulp3
|
tubby-like protein 3 |
| chr4_-_156059414 | 0.68 |
ENSMUST00000184348.1
|
Ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr2_-_152580300 | 0.67 |
ENSMUST00000053180.3
|
Defb19
|
defensin beta 19 |
| chr13_-_49147931 | 0.67 |
ENSMUST00000162581.1
ENSMUST00000110097.2 ENSMUST00000049265.8 ENSMUST00000035538.6 ENSMUST00000110096.1 ENSMUST00000091623.3 |
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr1_-_170927567 | 0.66 |
ENSMUST00000046322.7
ENSMUST00000159171.1 |
Fcrla
|
Fc receptor-like A |
| chr3_+_96576984 | 0.66 |
ENSMUST00000148290.1
|
Gm16253
|
predicted gene 16253 |
| chr4_+_134343466 | 0.65 |
ENSMUST00000105872.1
|
Slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr5_+_139405278 | 0.64 |
ENSMUST00000163940.1
|
C130050O18Rik
|
RIKEN cDNA C130050O18 gene |
| chr2_+_149830788 | 0.64 |
ENSMUST00000109935.1
|
Syndig1
|
synapse differentiation inducing 1 |
| chr7_-_30419802 | 0.63 |
ENSMUST00000075062.3
|
Hcst
|
hematopoietic cell signal transducer |
| chr12_-_76795489 | 0.63 |
ENSMUST00000082431.3
|
Gpx2
|
glutathione peroxidase 2 |
| chr17_+_50698525 | 0.62 |
ENSMUST00000061681.7
|
Gm7334
|
predicted gene 7334 |
| chr11_-_26591729 | 0.61 |
ENSMUST00000109504.1
|
Vrk2
|
vaccinia related kinase 2 |
| chr15_-_102366314 | 0.61 |
ENSMUST00000078508.5
|
Sp7
|
Sp7 transcription factor 7 |
| chr19_+_45363734 | 0.61 |
ENSMUST00000065601.5
ENSMUST00000111936.2 |
Btrc
|
beta-transducin repeat containing protein |
| chr17_+_35001282 | 0.61 |
ENSMUST00000174260.1
|
Vars
|
valyl-tRNA synthetase |
| chr19_-_5098418 | 0.60 |
ENSMUST00000025805.6
|
Cnih2
|
cornichon homolog 2 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hand1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 19.7 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 2.0 | 11.7 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 1.5 | 4.6 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 1.1 | 5.6 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 1.1 | 3.2 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 1.0 | 2.9 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.8 | 8.5 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.7 | 2.0 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.5 | 2.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.5 | 1.5 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.5 | 3.0 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.5 | 2.4 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.4 | 1.8 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 0.4 | 2.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.4 | 1.3 | GO:0071676 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.4 | 3.0 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.4 | 1.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 2.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.4 | 1.9 | GO:0050904 | diapedesis(GO:0050904) |
| 0.3 | 4.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.3 | 3.9 | GO:0035898 | parathyroid hormone secretion(GO:0035898) calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.3 | 0.9 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.3 | 0.9 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.3 | 0.9 | GO:0002588 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.3 | 1.6 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.3 | 1.0 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.3 | 0.8 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.3 | 2.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 0.2 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.2 | 1.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 3.3 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 1.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.2 | 0.7 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.2 | 2.0 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) |
| 0.2 | 4.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.1 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.2 | 0.6 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.2 | 2.8 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.2 | 0.8 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) |
| 0.2 | 0.8 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.2 | 0.8 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.2 | 0.6 | GO:1901608 | regulation of vesicle transport along microtubule(GO:1901608) |
| 0.2 | 1.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 0.9 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 0.5 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.2 | 1.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 3.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 0.3 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.2 | 1.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.2 | 0.5 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.2 | 0.2 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.2 | 0.6 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.2 | 0.8 | GO:0001692 | histamine metabolic process(GO:0001692) histidine catabolic process(GO:0006548) |
| 0.2 | 0.6 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 0.8 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.4 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 0.7 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 1.3 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 3.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.7 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 2.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.8 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.1 | 0.7 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.4 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 0.8 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 3.5 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 2.9 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.1 | 0.8 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.5 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.1 | 0.7 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 1.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.4 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) positive regulation of transforming growth factor beta3 production(GO:0032916) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 2.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.6 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 2.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 1.3 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.2 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.5 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.1 | 4.1 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 0.7 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 1.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.8 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.1 | 0.3 | GO:0090063 | regulation of microtubule nucleation(GO:0010968) positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.5 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 1.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.0 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.2 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.1 | 1.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.1 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 0.1 | 0.5 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.2 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 0.4 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 1.1 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 1.3 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.1 | 0.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.6 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 2.0 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.1 | 0.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.4 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 1.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 2.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.7 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.4 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 1.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 1.6 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.3 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.1 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 1.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 2.2 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.1 | 2.1 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 1.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.5 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.6 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.3 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.3 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.7 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 1.8 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.4 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 1.2 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.9 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 1.1 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 1.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 1.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 1.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.7 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.3 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 5.4 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.7 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 1.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.4 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.6 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.7 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.7 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 1.4 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 1.8 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.3 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 2.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.0 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.7 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.7 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.2 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.8 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.1 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.3 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.5 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.7 | 3.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.7 | 3.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.7 | 2.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.6 | 1.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.5 | 1.6 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.5 | 3.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.4 | 1.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.3 | 1.3 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.3 | 3.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 1.4 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 0.9 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 0.9 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 0.8 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 1.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 2.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 1.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 3.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 1.0 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.1 | 2.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.7 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 2.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.7 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 1.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 6.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 3.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 1.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 4.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 1.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 2.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.9 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 5.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 6.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.2 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 16.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 5.7 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 11.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.1 | 3.3 | GO:0004771 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 1.0 | 12.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.9 | 4.6 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.8 | 2.4 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.8 | 2.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.7 | 2.9 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.7 | 2.0 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.5 | 3.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 3.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.4 | 4.8 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.4 | 3.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 1.6 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.4 | 2.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.4 | 2.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.4 | 1.9 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.3 | 1.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.3 | 1.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.3 | 1.5 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 1.4 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.3 | 3.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.2 | 2.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 0.7 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.2 | 0.7 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.2 | 0.7 | GO:0070737 | protein-glycine ligase activity, elongating(GO:0070737) |
| 0.2 | 1.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 1.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 3.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 0.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 1.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 1.4 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 1.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 2.3 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.2 | 1.3 | GO:0048495 | GTPase inhibitor activity(GO:0005095) Roundabout binding(GO:0048495) |
| 0.2 | 0.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 1.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 0.6 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.5 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.1 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.0 | GO:0052851 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 1.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.8 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.4 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 1.5 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.3 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.1 | 0.3 | GO:0070735 | protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.1 | 0.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.3 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 1.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.5 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 4.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.9 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 2.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.0 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.7 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.2 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 0.5 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 1.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 1.7 | GO:0051537 | ferrous iron binding(GO:0008198) 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 2.6 | GO:0022839 | ion gated channel activity(GO:0022839) |
| 0.1 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 1.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 1.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 4.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 3.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 2.7 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 9.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 1.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 4.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 2.1 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 3.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 3.0 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.7 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 1.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.7 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 4.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 5.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 4.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.9 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 1.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 2.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 1.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 3.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 5.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 19.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 4.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 5.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 2.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 4.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 5.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 3.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 2.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 2.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 7.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |