Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Gsx2_Hoxd3_Vax1
Z-value: 0.82
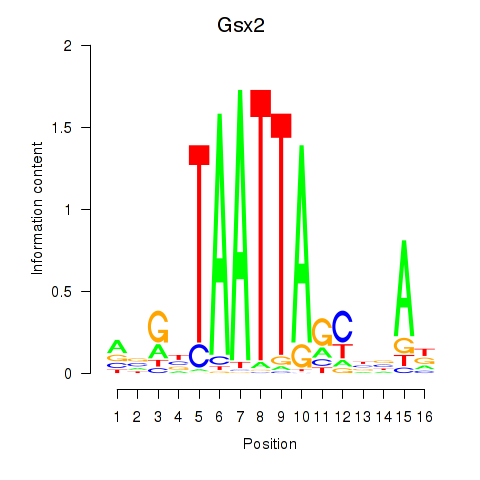
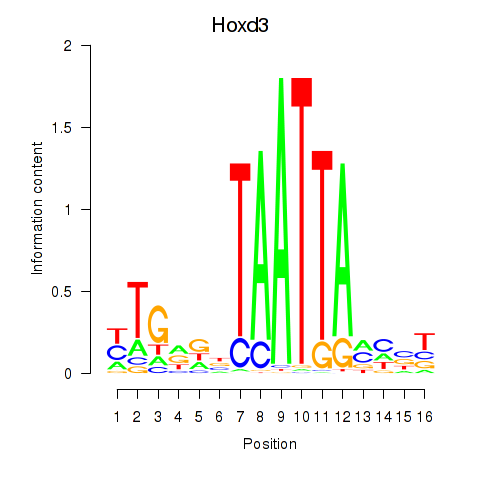
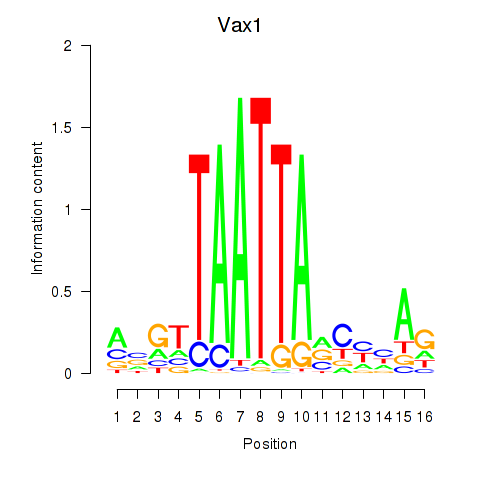
Transcription factors associated with Gsx2_Hoxd3_Vax1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gsx2
|
ENSMUSG00000035946.6 | GS homeobox 2 |
|
Hoxd3
|
ENSMUSG00000079277.3 | homeobox D3 |
|
Vax1
|
ENSMUSG00000006270.6 | ventral anterior homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vax1 | mm10_v2_chr19_-_59170978_59170978 | -0.57 | 2.8e-04 | Click! |
| Hoxd3 | mm10_v2_chr2_+_74711911_74711939 | -0.39 | 1.8e-02 | Click! |
Activity profile of Gsx2_Hoxd3_Vax1 motif
Sorted Z-values of Gsx2_Hoxd3_Vax1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_127898515 | 13.20 |
ENSMUST00000047134.7
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr1_+_130826762 | 9.21 |
ENSMUST00000133792.1
|
Pigr
|
polymeric immunoglobulin receptor |
| chr19_-_7966000 | 6.58 |
ENSMUST00000182102.1
ENSMUST00000075619.4 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr7_-_48843663 | 4.80 |
ENSMUST00000167786.2
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr2_+_109917639 | 3.77 |
ENSMUST00000046548.7
ENSMUST00000111037.2 |
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4 |
| chr2_+_116067213 | 3.72 |
ENSMUST00000152412.1
|
G630016G05Rik
|
RIKEN cDNA G630016G05 gene |
| chr18_+_56432116 | 3.24 |
ENSMUST00000070166.5
|
Gramd3
|
GRAM domain containing 3 |
| chr1_+_88211956 | 2.91 |
ENSMUST00000073049.6
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr15_-_101892916 | 2.70 |
ENSMUST00000100179.1
|
Krt76
|
keratin 76 |
| chr10_+_63024315 | 2.67 |
ENSMUST00000124784.1
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr18_-_75697639 | 2.66 |
ENSMUST00000165559.1
|
Ctif
|
CBP80/20-dependent translation initiation factor |
| chr17_+_85028347 | 2.37 |
ENSMUST00000024944.7
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr4_+_102589687 | 2.13 |
ENSMUST00000097949.4
ENSMUST00000106901.1 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chrX_-_143933089 | 2.07 |
ENSMUST00000087313.3
|
Dcx
|
doublecortin |
| chr3_-_131303144 | 1.94 |
ENSMUST00000106337.2
|
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chr4_+_133553370 | 1.91 |
ENSMUST00000042706.2
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr10_+_63024512 | 1.87 |
ENSMUST00000020262.4
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr19_-_8218832 | 1.82 |
ENSMUST00000113298.2
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr16_+_22918378 | 1.74 |
ENSMUST00000170805.1
|
Fetub
|
fetuin beta |
| chr4_-_14621669 | 1.72 |
ENSMUST00000143105.1
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr18_+_12741324 | 1.58 |
ENSMUST00000115857.2
ENSMUST00000121018.1 ENSMUST00000119108.1 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr10_+_128083273 | 1.57 |
ENSMUST00000026459.5
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr5_-_62766153 | 1.47 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr11_+_114851142 | 1.44 |
ENSMUST00000133245.1
ENSMUST00000122967.2 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_+_114851507 | 1.43 |
ENSMUST00000177952.1
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chrX_+_160768179 | 1.39 |
ENSMUST00000112368.2
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chrM_+_14138 | 1.36 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr12_-_98577940 | 1.35 |
ENSMUST00000110113.1
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chrX_+_160768013 | 1.35 |
ENSMUST00000033650.7
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chrX_+_107255878 | 1.35 |
ENSMUST00000101294.2
ENSMUST00000118820.1 ENSMUST00000120971.1 |
Gpr174
|
G protein-coupled receptor 174 |
| chr9_+_121950988 | 1.33 |
ENSMUST00000043011.7
|
Fam198a
|
family with sequence similarity 198, member A |
| chr6_-_130026954 | 1.33 |
ENSMUST00000074056.2
|
Klra6
|
killer cell lectin-like receptor, subfamily A, member 6 |
| chr8_+_36489191 | 1.32 |
ENSMUST00000171777.1
|
6430573F11Rik
|
RIKEN cDNA 6430573F11 gene |
| chr8_+_45658666 | 1.23 |
ENSMUST00000134675.1
ENSMUST00000139869.1 ENSMUST00000126067.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr14_-_64455903 | 1.22 |
ENSMUST00000067927.7
|
Msra
|
methionine sulfoxide reductase A |
| chr15_+_25773985 | 1.20 |
ENSMUST00000125667.1
|
Myo10
|
myosin X |
| chr3_+_138313279 | 1.18 |
ENSMUST00000013455.6
ENSMUST00000106247.1 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr15_-_101802343 | 1.17 |
ENSMUST00000063292.6
|
Krt73
|
keratin 73 |
| chr4_-_42661893 | 1.17 |
ENSMUST00000108006.3
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chrM_-_14060 | 1.15 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr5_+_87000838 | 1.11 |
ENSMUST00000031186.7
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr9_+_32116040 | 1.10 |
ENSMUST00000174641.1
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr2_+_69897220 | 1.09 |
ENSMUST00000055758.9
ENSMUST00000112251.2 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr5_+_87666200 | 1.08 |
ENSMUST00000094641.4
|
Csn1s1
|
casein alpha s1 |
| chr14_+_48446128 | 1.03 |
ENSMUST00000124720.1
|
Tmem260
|
transmembrane protein 260 |
| chr5_-_62765618 | 1.02 |
ENSMUST00000159470.1
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr11_-_99374895 | 1.02 |
ENSMUST00000006963.2
|
Krt28
|
keratin 28 |
| chrM_+_9870 | 0.94 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr1_+_88055377 | 0.93 |
ENSMUST00000138182.1
ENSMUST00000113142.3 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr11_-_49113757 | 0.93 |
ENSMUST00000060398.1
|
Olfr1396
|
olfactory receptor 1396 |
| chr1_+_88055467 | 0.92 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr4_-_14621494 | 0.91 |
ENSMUST00000149633.1
|
Slc26a7
|
solute carrier family 26, member 7 |
| chrM_+_10167 | 0.90 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr2_+_91457501 | 0.89 |
ENSMUST00000028689.3
|
Lrp4
|
low density lipoprotein receptor-related protein 4 |
| chr19_-_39649046 | 0.88 |
ENSMUST00000067328.6
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr9_-_97369958 | 0.84 |
ENSMUST00000035026.4
|
Trim42
|
tripartite motif-containing 42 |
| chr14_+_47298260 | 0.82 |
ENSMUST00000166743.1
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr4_-_129227883 | 0.82 |
ENSMUST00000106051.1
|
C77080
|
expressed sequence C77080 |
| chr4_+_100478806 | 0.81 |
ENSMUST00000133493.2
ENSMUST00000092730.3 ENSMUST00000106979.3 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr7_-_24724237 | 0.80 |
ENSMUST00000081657.4
|
Gm4763
|
predicted gene 4763 |
| chr8_+_45658731 | 0.80 |
ENSMUST00000143820.1
ENSMUST00000132139.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_-_125380793 | 0.78 |
ENSMUST00000042647.6
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr15_-_37459327 | 0.78 |
ENSMUST00000119730.1
ENSMUST00000120746.1 |
Ncald
|
neurocalcin delta |
| chr1_-_24612700 | 0.77 |
ENSMUST00000088336.1
|
Gm10222
|
predicted gene 10222 |
| chr6_-_130193112 | 0.76 |
ENSMUST00000112032.1
ENSMUST00000071554.2 |
Klra9
|
killer cell lectin-like receptor subfamily A, member 9 |
| chr13_-_17694729 | 0.75 |
ENSMUST00000068545.4
|
5033411D12Rik
|
RIKEN cDNA 5033411D12 gene |
| chr11_-_73326807 | 0.74 |
ENSMUST00000134079.1
|
Aspa
|
aspartoacylase |
| chr2_-_164613600 | 0.73 |
ENSMUST00000094351.4
ENSMUST00000109338.1 |
Wfdc8
|
WAP four-disulfide core domain 8 |
| chr2_+_164613519 | 0.71 |
ENSMUST00000094346.2
|
Wfdc6b
|
WAP four-disulfide core domain 6B |
| chr10_-_8886033 | 0.69 |
ENSMUST00000015449.5
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr14_-_100149764 | 0.69 |
ENSMUST00000097079.4
|
Klf12
|
Kruppel-like factor 12 |
| chr4_-_148152059 | 0.68 |
ENSMUST00000056965.5
ENSMUST00000168503.1 ENSMUST00000152098.1 |
Fbxo6
|
F-box protein 6 |
| chr2_+_69897255 | 0.68 |
ENSMUST00000131553.1
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr12_+_31184623 | 0.66 |
ENSMUST00000179160.1
|
6030469F06Rik
|
RIKEN cDNA 6030469F06 gene |
| chrX_-_143933204 | 0.66 |
ENSMUST00000112851.1
ENSMUST00000112856.2 ENSMUST00000033642.3 |
Dcx
|
doublecortin |
| chr1_-_141257158 | 0.66 |
ENSMUST00000179202.1
|
Gm4845
|
predicted gene 4845 |
| chr3_+_5218516 | 0.66 |
ENSMUST00000175866.1
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr6_-_136875794 | 0.66 |
ENSMUST00000032342.1
|
Mgp
|
matrix Gla protein |
| chr15_-_8710734 | 0.65 |
ENSMUST00000005493.7
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr4_+_95557494 | 0.65 |
ENSMUST00000079223.4
ENSMUST00000177394.1 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr5_-_84417359 | 0.64 |
ENSMUST00000113401.1
|
Epha5
|
Eph receptor A5 |
| chr13_+_19342154 | 0.63 |
ENSMUST00000103566.3
|
Tcrg-C4
|
T cell receptor gamma, constant 4 |
| chr6_+_37870786 | 0.63 |
ENSMUST00000120428.1
ENSMUST00000031859.7 |
Trim24
|
tripartite motif-containing 24 |
| chr16_-_45724600 | 0.63 |
ENSMUST00000096057.4
|
Tagln3
|
transgelin 3 |
| chr6_-_130129898 | 0.62 |
ENSMUST00000014476.5
|
Klra8
|
killer cell lectin-like receptor, subfamily A, member 8 |
| chr9_-_70934808 | 0.62 |
ENSMUST00000034731.8
|
Lipc
|
lipase, hepatic |
| chr4_+_95967205 | 0.62 |
ENSMUST00000030306.7
|
Hook1
|
hook homolog 1 (Drosophila) |
| chr1_+_75435930 | 0.61 |
ENSMUST00000037796.7
ENSMUST00000113584.1 ENSMUST00000145166.1 ENSMUST00000143730.1 ENSMUST00000133418.1 ENSMUST00000144874.1 ENSMUST00000140287.1 |
Gmppa
|
GDP-mannose pyrophosphorylase A |
| chr1_+_75436002 | 0.60 |
ENSMUST00000131545.1
ENSMUST00000141124.1 |
Gmppa
|
GDP-mannose pyrophosphorylase A |
| chr11_-_73326472 | 0.59 |
ENSMUST00000155630.2
|
Aspa
|
aspartoacylase |
| chr8_-_86580664 | 0.59 |
ENSMUST00000131423.1
ENSMUST00000152438.1 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr14_-_48662740 | 0.59 |
ENSMUST00000122009.1
|
Otx2
|
orthodenticle homolog 2 (Drosophila) |
| chr12_+_111814170 | 0.59 |
ENSMUST00000021714.7
|
Zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr11_+_116843278 | 0.59 |
ENSMUST00000106370.3
|
Mettl23
|
methyltransferase like 23 |
| chrX_-_99470672 | 0.58 |
ENSMUST00000113797.3
ENSMUST00000113790.1 ENSMUST00000036354.6 ENSMUST00000167246.1 |
Pja1
|
praja1, RING-H2 motif containing |
| chr15_-_82794236 | 0.57 |
ENSMUST00000006094.4
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26 |
| chr9_-_103222063 | 0.57 |
ENSMUST00000170904.1
|
Trf
|
transferrin |
| chr4_+_108719649 | 0.57 |
ENSMUST00000178992.1
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr7_-_14123042 | 0.56 |
ENSMUST00000098809.2
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr15_-_8710409 | 0.54 |
ENSMUST00000157065.1
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr5_-_88675190 | 0.54 |
ENSMUST00000133532.1
ENSMUST00000150438.1 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr4_-_15149755 | 0.52 |
ENSMUST00000108273.1
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr2_+_91257323 | 0.52 |
ENSMUST00000111349.2
ENSMUST00000131711.1 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr9_+_72958785 | 0.51 |
ENSMUST00000098567.2
ENSMUST00000034734.8 |
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 homolog (human) |
| chr13_-_53473074 | 0.50 |
ENSMUST00000021922.8
|
Msx2
|
msh homeobox 2 |
| chrM_+_7759 | 0.50 |
ENSMUST00000082407.1
ENSMUST00000082408.1 |
mt-Atp8
mt-Atp6
|
mitochondrially encoded ATP synthase 8 mitochondrially encoded ATP synthase 6 |
| chr14_+_56887795 | 0.49 |
ENSMUST00000022511.8
|
Zmym2
|
zinc finger, MYM-type 2 |
| chr9_+_110052016 | 0.49 |
ENSMUST00000164930.1
ENSMUST00000163979.1 |
Map4
|
microtubule-associated protein 4 |
| chr3_-_15426427 | 0.49 |
ENSMUST00000099201.3
|
Sirpb1a
|
signal-regulatory protein beta 1A |
| chr8_-_41016749 | 0.48 |
ENSMUST00000117735.1
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr11_+_75532099 | 0.48 |
ENSMUST00000169547.2
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr11_+_58171648 | 0.48 |
ENSMUST00000020820.1
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr10_-_83648631 | 0.48 |
ENSMUST00000146876.2
ENSMUST00000176294.1 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr2_-_28916412 | 0.47 |
ENSMUST00000050776.2
ENSMUST00000113849.1 |
Barhl1
|
BarH-like 1 (Drosophila) |
| chr16_-_32877723 | 0.47 |
ENSMUST00000119810.1
|
1700021K19Rik
|
RIKEN cDNA 1700021K19 gene |
| chr2_+_20737306 | 0.47 |
ENSMUST00000114606.1
ENSMUST00000114608.1 |
Etl4
|
enhancer trap locus 4 |
| chr13_-_56482246 | 0.46 |
ENSMUST00000022019.3
|
Il9
|
interleukin 9 |
| chr2_-_28916668 | 0.46 |
ENSMUST00000113847.1
|
Barhl1
|
BarH-like 1 (Drosophila) |
| chr7_+_38183217 | 0.45 |
ENSMUST00000165308.1
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr8_-_41041828 | 0.44 |
ENSMUST00000051379.7
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr9_-_55919605 | 0.44 |
ENSMUST00000037408.8
|
Scaper
|
S phase cyclin A-associated protein in the ER |
| chr8_-_13890233 | 0.44 |
ENSMUST00000033839.7
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr2_+_19371636 | 0.43 |
ENSMUST00000023856.8
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr6_-_149101674 | 0.42 |
ENSMUST00000111557.1
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr14_+_69347587 | 0.41 |
ENSMUST00000064831.5
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr10_+_128337761 | 0.40 |
ENSMUST00000005826.7
|
Cs
|
citrate synthase |
| chr16_-_16600533 | 0.40 |
ENSMUST00000159542.1
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr5_-_108795352 | 0.40 |
ENSMUST00000004943.1
|
Tmed11
|
transmembrane emp24 protein transport domain containing |
| chr19_-_39812744 | 0.40 |
ENSMUST00000162507.1
ENSMUST00000160476.1 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr5_-_3647806 | 0.39 |
ENSMUST00000119783.1
ENSMUST00000007559.8 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr15_+_41830921 | 0.39 |
ENSMUST00000166917.1
|
Oxr1
|
oxidation resistance 1 |
| chr12_+_80644212 | 0.38 |
ENSMUST00000085245.5
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr9_+_110476985 | 0.38 |
ENSMUST00000084948.4
ENSMUST00000061155.6 ENSMUST00000140686.1 ENSMUST00000084952.5 |
Kif9
|
kinesin family member 9 |
| chr13_-_102905740 | 0.38 |
ENSMUST00000167462.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_+_19368498 | 0.38 |
ENSMUST00000132655.1
|
Ppp1r13l
|
protein phosphatase 1, regulatory (inhibitor) subunit 13 like |
| chr14_+_53845234 | 0.38 |
ENSMUST00000103674.4
|
Trav19
|
T cell receptor alpha variable 19 |
| chr9_-_50659780 | 0.37 |
ENSMUST00000034567.3
|
Dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr6_+_8948608 | 0.37 |
ENSMUST00000160300.1
|
Nxph1
|
neurexophilin 1 |
| chr4_-_150914401 | 0.37 |
ENSMUST00000105675.1
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr5_+_104202609 | 0.37 |
ENSMUST00000066708.5
|
Dmp1
|
dentin matrix protein 1 |
| chr13_-_67332525 | 0.36 |
ENSMUST00000168892.1
ENSMUST00000109735.2 |
Zfp595
|
zinc finger protein 595 |
| chr8_-_121541939 | 0.35 |
ENSMUST00000034265.4
|
1700018B08Rik
|
RIKEN cDNA 1700018B08 gene |
| chr6_+_63255971 | 0.35 |
ENSMUST00000159561.1
ENSMUST00000095852.3 |
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr6_-_30693676 | 0.34 |
ENSMUST00000169422.1
ENSMUST00000115131.1 ENSMUST00000115130.2 ENSMUST00000031810.8 |
Cep41
|
centrosomal protein 41 |
| chr18_+_77332394 | 0.34 |
ENSMUST00000148341.1
|
Loxhd1
|
lipoxygenase homology domains 1 |
| chr12_-_84617326 | 0.34 |
ENSMUST00000021666.4
|
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr17_-_70853482 | 0.33 |
ENSMUST00000118283.1
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr13_-_49945335 | 0.33 |
ENSMUST00000178715.1
|
Gm10784
|
predicted pseudogene 10784 |
| chr5_-_63968867 | 0.32 |
ENSMUST00000154169.1
|
Rell1
|
RELT-like 1 |
| chr9_+_37208291 | 0.31 |
ENSMUST00000034632.8
|
Tmem218
|
transmembrane protein 218 |
| chrX_-_160138375 | 0.31 |
ENSMUST00000033662.8
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chr14_+_26693267 | 0.30 |
ENSMUST00000022433.4
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr11_+_59306920 | 0.30 |
ENSMUST00000000128.3
ENSMUST00000108783.3 |
Wnt9a
|
wingless-type MMTV integration site 9A |
| chr11_+_101087277 | 0.29 |
ENSMUST00000107302.1
ENSMUST00000107303.3 ENSMUST00000017945.8 ENSMUST00000149597.1 |
Mlx
|
MAX-like protein X |
| chr3_-_117077760 | 0.29 |
ENSMUST00000029642.5
|
1700061I17Rik
|
RIKEN cDNA 1700061I17 gene |
| chr15_+_39006272 | 0.29 |
ENSMUST00000179165.1
ENSMUST00000022906.7 |
Fzd6
|
frizzled homolog 6 (Drosophila) |
| chr4_+_145670685 | 0.28 |
ENSMUST00000105738.2
|
Gm13242
|
predicted gene 13242 |
| chr4_-_45532470 | 0.28 |
ENSMUST00000147448.1
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr9_+_27299205 | 0.28 |
ENSMUST00000115247.1
ENSMUST00000133213.1 |
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr16_-_97763712 | 0.28 |
ENSMUST00000019386.8
|
Ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chrX_+_37493428 | 0.27 |
ENSMUST00000115179.3
|
Rhox2d
|
reproductive homeobox 2D |
| chr12_+_87874070 | 0.27 |
ENSMUST00000110147.2
|
Gm2016
|
predicted gene 2016 |
| chr9_+_94669876 | 0.27 |
ENSMUST00000033463.9
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr14_+_64589802 | 0.27 |
ENSMUST00000180610.1
|
A930011O12Rik
|
RIKEN cDNA A930011O12 gene |
| chr18_+_33464163 | 0.27 |
ENSMUST00000097634.3
|
Gm10549
|
predicted gene 10549 |
| chr1_-_163725123 | 0.26 |
ENSMUST00000159679.1
|
Mettl11b
|
methyltransferase like 11B |
| chr16_-_92400067 | 0.25 |
ENSMUST00000023672.8
|
Rcan1
|
regulator of calcineurin 1 |
| chr7_-_8161654 | 0.25 |
ENSMUST00000168807.2
|
Vmn2r41
|
vomeronasal 2, receptor 41 |
| chr11_+_73371246 | 0.24 |
ENSMUST00000120401.1
ENSMUST00000078952.2 ENSMUST00000170592.1 ENSMUST00000127789.1 |
Olfr376
|
olfactory receptor 376 |
| chr3_+_66219909 | 0.24 |
ENSMUST00000029421.5
|
Ptx3
|
pentraxin related gene |
| chr8_-_109251698 | 0.23 |
ENSMUST00000079189.3
|
4922502B01Rik
|
RIKEN cDNA 4922502B01 gene |
| chr9_-_110476637 | 0.23 |
ENSMUST00000111934.1
ENSMUST00000068025.6 |
Klhl18
|
kelch-like 18 |
| chr5_-_136986829 | 0.22 |
ENSMUST00000034953.7
ENSMUST00000085941.5 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr1_+_88306731 | 0.22 |
ENSMUST00000040210.7
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chrM_+_8600 | 0.21 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr3_+_19187321 | 0.21 |
ENSMUST00000130806.1
ENSMUST00000117529.1 ENSMUST00000119865.1 |
Mtfr1
|
mitochondrial fission regulator 1 |
| chr1_-_161070613 | 0.20 |
ENSMUST00000035430.3
|
Dars2
|
aspartyl-tRNA synthetase 2 (mitochondrial) |
| chr18_+_37355271 | 0.20 |
ENSMUST00000051163.1
|
Pcdhb8
|
protocadherin beta 8 |
| chrX_+_37530452 | 0.19 |
ENSMUST00000072167.3
ENSMUST00000184746.1 |
Rhox2e
|
reproductive homeobox 2E |
| chr5_-_106926245 | 0.19 |
ENSMUST00000117588.1
|
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr6_+_134640940 | 0.19 |
ENSMUST00000062755.8
|
Loh12cr1
|
loss of heterozygosity, 12, chromosomal region 1 homolog (human) |
| chrX_+_37412067 | 0.19 |
ENSMUST00000115191.4
|
Rhox2b
|
reproductive homeobox 2B |
| chrX_+_37244976 | 0.18 |
ENSMUST00000063340.5
ENSMUST00000173650.1 |
Rhox2a
|
reproductive homeobox 2A |
| chr16_+_32877775 | 0.18 |
ENSMUST00000023489.4
ENSMUST00000171325.1 |
Fyttd1
|
forty-two-three domain containing 1 |
| chr7_-_9223653 | 0.17 |
ENSMUST00000072787.4
|
Vmn2r37
|
vomeronasal 2, receptor 37 |
| chr8_+_21839926 | 0.17 |
ENSMUST00000006745.3
|
Defb2
|
defensin beta 2 |
| chr16_-_50330987 | 0.16 |
ENSMUST00000114488.1
|
Bbx
|
bobby sox homolog (Drosophila) |
| chr11_+_90883320 | 0.16 |
ENSMUST00000142163.1
ENSMUST00000132218.1 |
4930405D11Rik
|
RIKEN cDNA 4930405D11 gene |
| chr3_+_125404292 | 0.16 |
ENSMUST00000144344.1
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr9_-_85749308 | 0.15 |
ENSMUST00000039213.8
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr7_-_7689513 | 0.15 |
ENSMUST00000173459.1
|
Vmn2r34
|
vomeronasal 2, receptor 34 |
| chr3_+_53488677 | 0.15 |
ENSMUST00000029307.3
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chr18_+_52615908 | 0.14 |
ENSMUST00000072666.3
|
Zfp474
|
zinc finger protein 474 |
| chr4_-_14621805 | 0.14 |
ENSMUST00000042221.7
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr1_-_83038431 | 0.14 |
ENSMUST00000164473.1
ENSMUST00000045560.8 |
Slc19a3
|
solute carrier family 19, member 3 |
| chr3_-_33082004 | 0.14 |
ENSMUST00000108225.3
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr1_-_33814516 | 0.13 |
ENSMUST00000044455.5
ENSMUST00000115167.1 |
Zfp451
|
zinc finger protein 451 |
| chr18_+_57142782 | 0.13 |
ENSMUST00000139892.1
|
Megf10
|
multiple EGF-like-domains 10 |
| chr3_+_92992213 | 0.13 |
ENSMUST00000090863.3
|
Lce3f
|
late cornified envelope 3F |
| chr3_-_64565298 | 0.13 |
ENSMUST00000176481.1
|
Vmn2r6
|
vomeronasal 2, receptor 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gsx2_Hoxd3_Vax1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 1.6 | 4.8 | GO:1903919 | regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) |
| 1.3 | 3.8 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 1.0 | 2.9 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.7 | 2.7 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.6 | 8.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.4 | 1.8 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 2.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.3 | 1.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.3 | 0.8 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.2 | 0.7 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 1.4 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.2 | 1.6 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.2 | 1.3 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.2 | 0.9 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.2 | 2.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.2 | 1.9 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 2.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 2.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.4 | GO:1903189 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 1.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.6 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.6 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.7 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.4 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 1.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.6 | GO:0097460 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 2.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.6 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.1 | 0.6 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 0.8 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 1.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.4 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.0 | 0.6 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 1.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 | 0.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.4 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.3 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.5 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 1.3 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 1.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.9 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 2.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) recognition of apoptotic cell(GO:0043654) |
| 0.0 | 1.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.5 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 14.0 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.0 | 0.6 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.4 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 1.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.8 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 8.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 1.6 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.4 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 0.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 1.4 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 3.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 2.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.9 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 3.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 3.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 4.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 16.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.9 | 9.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.8 | 4.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.6 | 13.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.6 | 8.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.4 | 1.3 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.4 | 1.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.3 | 1.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.2 | 2.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 3.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 1.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 0.6 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.2 | 1.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 1.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 2.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 5.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.7 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.4 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 1.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 3.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.6 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.6 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.6 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.1 | 2.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.4 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 2.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.9 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 2.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 4.6 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 9.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.5 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 1.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 2.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 2.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.7 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |