Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
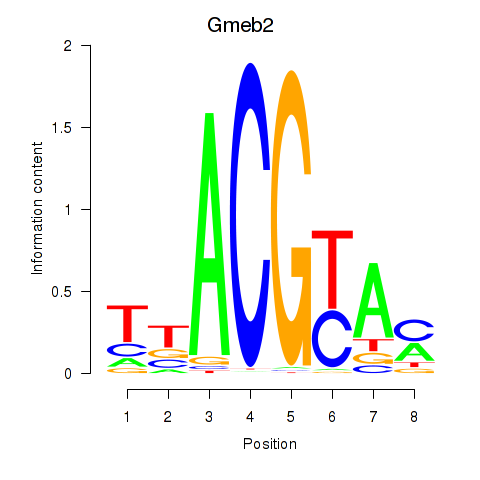
Results for Gmeb2
Z-value: 0.88
Transcription factors associated with Gmeb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gmeb2
|
ENSMUSG00000038705.7 | glucocorticoid modulatory element binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gmeb2 | mm10_v2_chr2_-_181288016_181288041 | 0.65 | 1.6e-05 | Click! |
Activity profile of Gmeb2 motif
Sorted Z-values of Gmeb2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_79660196 | 5.43 |
ENSMUST00000035977.7
|
Ticrr
|
TOPBP1-interacting checkpoint and replication regulator |
| chrX_-_136068236 | 5.07 |
ENSMUST00000049130.7
|
Bex2
|
brain expressed X-linked 2 |
| chr1_-_88205674 | 4.47 |
ENSMUST00000119972.2
|
Dnajb3
|
DnaJ (Hsp40) homolog, subfamily B, member 3 |
| chr8_-_92356103 | 4.46 |
ENSMUST00000034183.3
|
4933436C20Rik
|
RIKEN cDNA 4933436C20 gene |
| chr2_-_26021679 | 3.72 |
ENSMUST00000036509.7
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr2_-_26021532 | 3.64 |
ENSMUST00000136750.1
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr11_+_78301529 | 3.50 |
ENSMUST00000045026.3
|
Spag5
|
sperm associated antigen 5 |
| chr8_-_92355764 | 3.20 |
ENSMUST00000180102.1
ENSMUST00000179421.1 ENSMUST00000179222.1 ENSMUST00000179029.1 |
4933436C20Rik
|
RIKEN cDNA 4933436C20 gene |
| chr6_-_70792155 | 3.17 |
ENSMUST00000066134.5
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr15_-_99651580 | 3.06 |
ENSMUST00000171908.1
ENSMUST00000171702.1 ENSMUST00000109581.2 ENSMUST00000169810.1 ENSMUST00000023756.5 |
Racgap1
|
Rac GTPase-activating protein 1 |
| chr12_+_102554966 | 2.92 |
ENSMUST00000021610.5
|
Chga
|
chromogranin A |
| chr11_+_116198853 | 2.85 |
ENSMUST00000021130.6
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr6_-_112696604 | 2.74 |
ENSMUST00000113182.1
ENSMUST00000113180.1 ENSMUST00000068487.5 ENSMUST00000077088.4 |
Rad18
|
RAD18 homolog (S. cerevisiae) |
| chr12_-_110978981 | 2.72 |
ENSMUST00000135131.1
ENSMUST00000043459.6 ENSMUST00000128353.1 |
Ankrd9
|
ankyrin repeat domain 9 |
| chr2_+_109280738 | 2.67 |
ENSMUST00000028527.7
|
Kif18a
|
kinesin family member 18A |
| chr3_+_127553462 | 2.65 |
ENSMUST00000043108.4
|
4930422G04Rik
|
RIKEN cDNA 4930422G04 gene |
| chr12_-_110978618 | 2.62 |
ENSMUST00000140788.1
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr18_-_62179948 | 2.58 |
ENSMUST00000053640.3
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr12_-_110978943 | 2.56 |
ENSMUST00000142012.1
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr11_-_40733373 | 2.46 |
ENSMUST00000020579.8
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr13_-_49652714 | 2.32 |
ENSMUST00000021818.7
|
Cenpp
|
centromere protein P |
| chr1_+_175880775 | 2.18 |
ENSMUST00000039725.6
|
Exo1
|
exonuclease 1 |
| chr2_-_18998126 | 2.05 |
ENSMUST00000006912.5
|
Pip4k2a
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr10_+_58446845 | 2.00 |
ENSMUST00000003310.5
|
Ranbp2
|
RAN binding protein 2 |
| chr2_+_18998332 | 2.00 |
ENSMUST00000028069.1
|
4930426L09Rik
|
RIKEN cDNA 4930426L09 gene |
| chr7_+_102441685 | 1.92 |
ENSMUST00000033283.9
|
Rrm1
|
ribonucleotide reductase M1 |
| chr5_+_143548700 | 1.92 |
ENSMUST00000169329.1
ENSMUST00000067145.5 ENSMUST00000119488.1 ENSMUST00000118121.1 |
Fam220a
Fam220a
|
family with sequence similarity 220, member A family with sequence similarity 220, member A |
| chr6_-_67535783 | 1.91 |
ENSMUST00000058178.4
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr19_-_40588453 | 1.90 |
ENSMUST00000025979.6
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr9_-_64172879 | 1.87 |
ENSMUST00000176299.1
ENSMUST00000130127.1 ENSMUST00000176794.1 ENSMUST00000177045.1 |
Zwilch
|
zwilch kinetochore protein |
| chr19_-_40588374 | 1.83 |
ENSMUST00000175932.1
ENSMUST00000176955.1 ENSMUST00000149476.2 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr19_-_40588338 | 1.79 |
ENSMUST00000176939.1
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr11_-_109472611 | 1.75 |
ENSMUST00000168740.1
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr1_-_162740540 | 1.73 |
ENSMUST00000028016.9
ENSMUST00000182660.1 |
Prrc2c
|
proline-rich coiled-coil 2C |
| chr6_-_30958990 | 1.65 |
ENSMUST00000101589.3
|
Klf14
|
Kruppel-like factor 14 |
| chr13_+_81657732 | 1.62 |
ENSMUST00000049055.6
|
Lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr1_-_162740350 | 1.60 |
ENSMUST00000182331.1
ENSMUST00000183011.1 ENSMUST00000182593.1 ENSMUST00000182149.1 |
Prrc2c
|
proline-rich coiled-coil 2C |
| chr4_-_4138432 | 1.59 |
ENSMUST00000070375.7
|
Penk
|
preproenkephalin |
| chr7_-_4546567 | 1.57 |
ENSMUST00000065957.5
|
Syt5
|
synaptotagmin V |
| chr15_+_82016420 | 1.53 |
ENSMUST00000168581.1
ENSMUST00000170630.1 ENSMUST00000164779.1 |
Xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr17_+_47593516 | 1.48 |
ENSMUST00000182874.1
|
Ccnd3
|
cyclin D3 |
| chr12_+_17348422 | 1.48 |
ENSMUST00000046011.10
|
Nol10
|
nucleolar protein 10 |
| chr3_+_131110350 | 1.48 |
ENSMUST00000066849.6
ENSMUST00000106341.2 ENSMUST00000029611.7 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr11_+_69095217 | 1.44 |
ENSMUST00000101004.2
|
Per1
|
period circadian clock 1 |
| chr7_-_83884289 | 1.44 |
ENSMUST00000094216.3
|
Mesdc1
|
mesoderm development candidate 1 |
| chr9_+_119937606 | 1.44 |
ENSMUST00000035100.5
|
Ttc21a
|
tetratricopeptide repeat domain 21A |
| chr1_-_55027473 | 1.40 |
ENSMUST00000027127.7
|
Sf3b1
|
splicing factor 3b, subunit 1 |
| chr17_+_47593444 | 1.34 |
ENSMUST00000182209.1
|
Ccnd3
|
cyclin D3 |
| chr2_+_153741274 | 1.31 |
ENSMUST00000028981.8
|
Mapre1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr16_-_91044473 | 1.30 |
ENSMUST00000118522.1
|
Paxbp1
|
PAX3 and PAX7 binding protein 1 |
| chr12_+_33429605 | 1.28 |
ENSMUST00000020877.7
|
Twistnb
|
TWIST neighbor |
| chr2_-_37703845 | 1.24 |
ENSMUST00000155237.1
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr12_+_17544873 | 1.24 |
ENSMUST00000171737.1
|
Odc1
|
ornithine decarboxylase, structural 1 |
| chr6_-_29212240 | 1.24 |
ENSMUST00000160878.1
ENSMUST00000078155.5 |
Impdh1
|
inosine 5'-phosphate dehydrogenase 1 |
| chr2_-_163645125 | 1.23 |
ENSMUST00000017851.3
|
Serinc3
|
serine incorporator 3 |
| chr9_+_56418624 | 1.20 |
ENSMUST00000034879.3
|
Hmg20a
|
high mobility group 20A |
| chr7_+_66109474 | 1.19 |
ENSMUST00000036372.6
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chr11_-_85139939 | 1.15 |
ENSMUST00000108075.2
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr9_-_123717576 | 1.15 |
ENSMUST00000026274.7
|
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr4_-_4138817 | 1.12 |
ENSMUST00000133567.1
|
Penk
|
preproenkephalin |
| chr19_+_8967031 | 1.10 |
ENSMUST00000052248.7
|
Eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr1_+_52008210 | 1.10 |
ENSMUST00000027277.5
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr16_+_14361552 | 1.09 |
ENSMUST00000100167.3
ENSMUST00000154748.1 ENSMUST00000134776.1 |
Abcc1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
| chr5_+_146833106 | 1.01 |
ENSMUST00000075453.2
ENSMUST00000099272.2 |
Rpl21
|
ribosomal protein L21 |
| chr8_-_123949201 | 0.99 |
ENSMUST00000044795.7
|
Nup133
|
nucleoporin 133 |
| chr17_+_35001282 | 0.98 |
ENSMUST00000174260.1
|
Vars
|
valyl-tRNA synthetase |
| chr3_-_130730310 | 0.98 |
ENSMUST00000062601.7
|
Rpl34
|
ribosomal protein L34 |
| chrX_+_134601271 | 0.96 |
ENSMUST00000050331.6
ENSMUST00000059297.5 |
Hnrnph2
|
heterogeneous nuclear ribonucleoprotein H2 |
| chr2_+_125673077 | 0.95 |
ENSMUST00000164756.2
|
Eid1
|
EP300 interacting inhibitor of differentiation 1 |
| chrX_+_134601179 | 0.92 |
ENSMUST00000074950.4
ENSMUST00000113203.1 ENSMUST00000113202.1 |
Hnrnph2
|
heterogeneous nuclear ribonucleoprotein H2 |
| chr3_-_95217690 | 0.91 |
ENSMUST00000107209.1
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr5_+_146832890 | 0.91 |
ENSMUST00000035983.5
|
Rpl21
|
ribosomal protein L21 |
| chr15_-_58823530 | 0.90 |
ENSMUST00000072113.5
|
Tmem65
|
transmembrane protein 65 |
| chr1_-_46854046 | 0.90 |
ENSMUST00000027131.4
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr11_+_98809787 | 0.87 |
ENSMUST00000169695.1
|
Casc3
|
cancer susceptibility candidate 3 |
| chr19_-_4615453 | 0.82 |
ENSMUST00000053597.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr15_-_100425050 | 0.80 |
ENSMUST00000123461.1
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr15_+_82016369 | 0.79 |
ENSMUST00000069530.6
|
Xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr15_-_50889691 | 0.78 |
ENSMUST00000165201.2
ENSMUST00000184458.1 |
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr2_+_69723071 | 0.77 |
ENSMUST00000040915.8
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G) |
| chr9_+_64173364 | 0.77 |
ENSMUST00000034966.7
|
Rpl4
|
ribosomal protein L4 |
| chr2_-_77946180 | 0.77 |
ENSMUST00000111824.1
ENSMUST00000111819.1 ENSMUST00000128963.1 |
Cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr2_-_53191214 | 0.76 |
ENSMUST00000076313.6
ENSMUST00000125243.1 |
Prpf40a
|
PRP40 pre-mRNA processing factor 40 homolog A (yeast) |
| chr11_+_93886157 | 0.76 |
ENSMUST00000063718.4
ENSMUST00000107854.2 |
Mbtd1
|
mbt domain containing 1 |
| chr14_-_37135310 | 0.75 |
ENSMUST00000165649.2
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr6_+_51470339 | 0.75 |
ENSMUST00000094623.3
|
Cbx3
|
chromobox 3 |
| chrX_+_151522352 | 0.73 |
ENSMUST00000148622.1
|
Phf8
|
PHD finger protein 8 |
| chrX_+_139480365 | 0.73 |
ENSMUST00000046763.6
ENSMUST00000113030.2 |
D330045A20Rik
|
RIKEN cDNA D330045A20 gene |
| chr6_-_39725193 | 0.71 |
ENSMUST00000101497.3
|
Braf
|
Braf transforming gene |
| chr11_-_107348130 | 0.70 |
ENSMUST00000134763.1
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr3_-_95217741 | 0.69 |
ENSMUST00000107204.1
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr3_-_95217877 | 0.69 |
ENSMUST00000136139.1
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr2_+_162931520 | 0.68 |
ENSMUST00000130411.1
|
Srsf6
|
serine/arginine-rich splicing factor 6 |
| chrX_+_7841800 | 0.68 |
ENSMUST00000033494.9
ENSMUST00000115666.1 |
Otud5
|
OTU domain containing 5 |
| chr2_+_69722797 | 0.66 |
ENSMUST00000090858.3
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G) |
| chr18_-_43477764 | 0.66 |
ENSMUST00000057110.9
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr17_-_94749874 | 0.65 |
ENSMUST00000171284.1
|
Mettl4
|
methyltransferase like 4 |
| chr2_-_77946331 | 0.65 |
ENSMUST00000111821.2
ENSMUST00000111818.1 |
Cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr17_+_47140942 | 0.63 |
ENSMUST00000077951.7
|
Trerf1
|
transcriptional regulating factor 1 |
| chr4_-_34882919 | 0.61 |
ENSMUST00000098163.2
ENSMUST00000047950.5 |
Zfp292
|
zinc finger protein 292 |
| chr5_-_36695969 | 0.61 |
ENSMUST00000031091.9
ENSMUST00000140063.1 |
D5Ertd579e
|
DNA segment, Chr 5, ERATO Doi 579, expressed |
| chr11_+_58963790 | 0.61 |
ENSMUST00000075141.6
|
Trim17
|
tripartite motif-containing 17 |
| chr13_-_96670838 | 0.61 |
ENSMUST00000022176.8
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr4_+_44756609 | 0.60 |
ENSMUST00000143385.1
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr2_-_77946375 | 0.58 |
ENSMUST00000065889.3
|
Cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr5_+_86804214 | 0.58 |
ENSMUST00000119339.1
ENSMUST00000120498.1 |
Ythdc1
|
YTH domain containing 1 |
| chr2_+_105126505 | 0.56 |
ENSMUST00000143043.1
|
Wt1
|
Wilms tumor 1 homolog |
| chr5_+_29378604 | 0.56 |
ENSMUST00000181005.1
|
4632411P08Rik
|
RIKEN cDNA 4632411P08 gene |
| chr6_-_39725448 | 0.54 |
ENSMUST00000002487.8
|
Braf
|
Braf transforming gene |
| chr9_+_110132015 | 0.54 |
ENSMUST00000088716.5
ENSMUST00000111969.1 ENSMUST00000035057.7 ENSMUST00000111966.1 ENSMUST00000111968.1 |
Smarcc1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr13_-_17805093 | 0.53 |
ENSMUST00000042365.7
|
Cdk13
|
cyclin-dependent kinase 13 |
| chr13_+_49653297 | 0.53 |
ENSMUST00000021824.7
|
Nol8
|
nucleolar protein 8 |
| chr11_-_97782377 | 0.50 |
ENSMUST00000128801.1
|
Rpl23
|
ribosomal protein L23 |
| chr11_-_93885752 | 0.49 |
ENSMUST00000066888.3
|
Utp18
|
UTP18, small subunit (SSU) processome component, homolog (yeast) |
| chr11_-_97782409 | 0.48 |
ENSMUST00000103146.4
|
Rpl23
|
ribosomal protein L23 |
| chr6_-_51469869 | 0.45 |
ENSMUST00000114459.1
ENSMUST00000069949.6 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr1_-_91459254 | 0.45 |
ENSMUST00000069620.8
|
Per2
|
period circadian clock 2 |
| chr6_-_51469836 | 0.44 |
ENSMUST00000090002.7
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr2_-_140066661 | 0.42 |
ENSMUST00000046656.2
ENSMUST00000099304.3 ENSMUST00000110079.2 |
Tasp1
|
taspase, threonine aspartase 1 |
| chr9_-_59486323 | 0.42 |
ENSMUST00000165322.1
|
Arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila) |
| chrX_-_150588071 | 0.41 |
ENSMUST00000140207.1
ENSMUST00000112719.1 ENSMUST00000112727.3 ENSMUST00000112721.3 ENSMUST00000026303.9 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chrX_+_7842056 | 0.40 |
ENSMUST00000115667.3
ENSMUST00000115668.3 ENSMUST00000115665.1 |
Otud5
|
OTU domain containing 5 |
| chr1_-_55088156 | 0.40 |
ENSMUST00000127861.1
ENSMUST00000144077.1 |
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr9_+_59291565 | 0.39 |
ENSMUST00000026266.7
|
Adpgk
|
ADP-dependent glucokinase |
| chr11_+_23665615 | 0.38 |
ENSMUST00000109525.1
ENSMUST00000020520.4 |
Pus10
|
pseudouridylate synthase 10 |
| chr4_+_44756553 | 0.38 |
ENSMUST00000107824.2
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr15_-_78544345 | 0.36 |
ENSMUST00000053239.2
|
Sstr3
|
somatostatin receptor 3 |
| chr16_-_91646906 | 0.36 |
ENSMUST00000120450.1
ENSMUST00000023684.7 |
Gart
|
phosphoribosylglycinamide formyltransferase |
| chr17_+_86963077 | 0.36 |
ENSMUST00000024956.8
|
Rhoq
|
ras homolog gene family, member Q |
| chrX_+_13280970 | 0.35 |
ENSMUST00000000804.6
|
Ddx3x
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked |
| chr2_+_53192067 | 0.35 |
ENSMUST00000028336.6
|
Arl6ip6
|
ADP-ribosylation factor-like 6 interacting protein 6 |
| chr17_+_24414640 | 0.33 |
ENSMUST00000115371.1
ENSMUST00000088512.5 ENSMUST00000163717.1 |
Rnps1
|
ribonucleic acid binding protein S1 |
| chr7_-_45466894 | 0.32 |
ENSMUST00000033093.8
|
Bax
|
BCL2-associated X protein |
| chr11_-_84513485 | 0.31 |
ENSMUST00000018841.2
|
Aatf
|
apoptosis antagonizing transcription factor |
| chr8_-_124949165 | 0.31 |
ENSMUST00000034469.5
|
Egln1
|
EGL nine homolog 1 (C. elegans) |
| chr7_+_105736565 | 0.29 |
ENSMUST00000033182.3
|
Ilk
|
integrin linked kinase |
| chr7_+_29170345 | 0.29 |
ENSMUST00000033886.7
|
Ggn
|
gametogenetin |
| chr4_-_109665249 | 0.28 |
ENSMUST00000063531.4
|
Cdkn2c
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
| chr5_+_76183880 | 0.27 |
ENSMUST00000031144.7
|
Tmem165
|
transmembrane protein 165 |
| chr4_+_62408770 | 0.27 |
ENSMUST00000084524.3
|
Prpf4
|
PRP4 pre-mRNA processing factor 4 homolog (yeast) |
| chr7_-_55962466 | 0.25 |
ENSMUST00000032635.7
ENSMUST00000152649.1 |
Nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 homolog (human) |
| chr1_+_166379097 | 0.24 |
ENSMUST00000027846.7
|
Tada1
|
transcriptional adaptor 1 |
| chr1_+_143739016 | 0.24 |
ENSMUST00000145969.1
|
Glrx2
|
glutaredoxin 2 (thioltransferase) |
| chr11_+_40733639 | 0.24 |
ENSMUST00000020578.4
|
Nudcd2
|
NudC domain containing 2 |
| chr6_+_21949571 | 0.23 |
ENSMUST00000031680.3
ENSMUST00000115389.1 ENSMUST00000151473.1 |
Ing3
|
inhibitor of growth family, member 3 |
| chr8_+_25720054 | 0.22 |
ENSMUST00000068916.8
ENSMUST00000139836.1 |
Ppapdc1b
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr5_-_145201829 | 0.22 |
ENSMUST00000162220.1
ENSMUST00000031632.8 |
Zkscan14
|
zinc finger with KRAB and SCAN domains 14 |
| chr10_+_42583787 | 0.22 |
ENSMUST00000105497.1
ENSMUST00000144806.1 |
Ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr5_-_67099235 | 0.21 |
ENSMUST00000012664.8
|
Phox2b
|
paired-like homeobox 2b |
| chr17_+_85090647 | 0.21 |
ENSMUST00000095188.5
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr12_-_31654767 | 0.20 |
ENSMUST00000020977.2
|
Dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr2_-_36136773 | 0.19 |
ENSMUST00000028251.3
|
Rbm18
|
RNA binding motif protein 18 |
| chr1_-_55088024 | 0.19 |
ENSMUST00000027123.8
|
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr16_+_57549232 | 0.19 |
ENSMUST00000159414.1
|
Filip1l
|
filamin A interacting protein 1-like |
| chr7_-_111082997 | 0.19 |
ENSMUST00000161051.1
ENSMUST00000160132.1 ENSMUST00000106666.3 ENSMUST00000162415.1 |
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr12_-_56345862 | 0.18 |
ENSMUST00000021416.7
|
Mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr11_+_83302817 | 0.18 |
ENSMUST00000142680.1
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr4_-_40269778 | 0.18 |
ENSMUST00000042575.6
|
Topors
|
topoisomerase I binding, arginine/serine-rich |
| chr1_-_179517992 | 0.17 |
ENSMUST00000128302.1
ENSMUST00000111134.1 |
Smyd3
|
SET and MYND domain containing 3 |
| chr1_-_77515048 | 0.16 |
ENSMUST00000027451.6
|
Epha4
|
Eph receptor A4 |
| chr19_-_24477356 | 0.15 |
ENSMUST00000099556.1
|
Fam122a
|
family with sequence similarity 122, member A |
| chr7_-_55962424 | 0.14 |
ENSMUST00000126604.1
ENSMUST00000117812.1 ENSMUST00000119201.1 |
Nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 homolog (human) |
| chr8_+_106210936 | 0.14 |
ENSMUST00000071592.5
|
Prmt7
|
protein arginine N-methyltransferase 7 |
| chr8_+_106211016 | 0.14 |
ENSMUST00000109297.1
|
Prmt7
|
protein arginine N-methyltransferase 7 |
| chr15_-_79546741 | 0.13 |
ENSMUST00000054014.7
|
Ddx17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 |
| chr3_-_123236134 | 0.13 |
ENSMUST00000106427.1
ENSMUST00000106426.1 ENSMUST00000051443.5 |
Synpo2
|
synaptopodin 2 |
| chr11_-_102185239 | 0.12 |
ENSMUST00000021297.5
|
Lsm12
|
LSM12 homolog (S. cerevisiae) |
| chr12_-_59219725 | 0.12 |
ENSMUST00000043204.7
|
Fbxo33
|
F-box protein 33 |
| chr7_+_16944645 | 0.12 |
ENSMUST00000094807.5
|
Pnmal2
|
PNMA-like 2 |
| chr7_+_4137032 | 0.11 |
ENSMUST00000128756.1
ENSMUST00000132086.1 ENSMUST00000037472.6 ENSMUST00000117274.1 ENSMUST00000121270.1 |
Leng8
|
leukocyte receptor cluster (LRC) member 8 |
| chr11_-_116274102 | 0.10 |
ENSMUST00000106425.3
|
Srp68
|
signal recognition particle 68 |
| chr7_+_4915202 | 0.10 |
ENSMUST00000116354.2
|
Zfp628
|
zinc finger protein 628 |
| chr7_+_29170204 | 0.10 |
ENSMUST00000098609.2
|
Ggn
|
gametogenetin |
| chr11_-_69681822 | 0.10 |
ENSMUST00000005336.2
|
Senp3
|
SUMO/sentrin specific peptidase 3 |
| chr14_-_52104015 | 0.10 |
ENSMUST00000111610.4
ENSMUST00000164655.1 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr2_+_31670714 | 0.08 |
ENSMUST00000038474.7
ENSMUST00000137156.1 |
Exosc2
|
exosome component 2 |
| chr11_-_116274197 | 0.07 |
ENSMUST00000021133.9
|
Srp68
|
signal recognition particle 68 |
| chr12_+_111039334 | 0.07 |
ENSMUST00000084968.7
|
Rcor1
|
REST corepressor 1 |
| chr4_+_84884276 | 0.07 |
ENSMUST00000047023.6
|
Cntln
|
centlein, centrosomal protein |
| chr7_-_35802968 | 0.06 |
ENSMUST00000061586.4
|
Zfp507
|
zinc finger protein 507 |
| chr4_+_84884418 | 0.05 |
ENSMUST00000169371.2
|
Cntln
|
centlein, centrosomal protein |
| chr11_+_6200029 | 0.03 |
ENSMUST00000181545.1
|
A730071L15Rik
|
RIKEN cDNA A730071L15Rik gene |
| chr10_-_4387436 | 0.03 |
ENSMUST00000100077.3
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr1_+_185454803 | 0.01 |
ENSMUST00000061093.6
|
Slc30a10
|
solute carrier family 30, member 10 |
| chr11_-_69681903 | 0.01 |
ENSMUST00000066760.1
|
Senp3
|
SUMO/sentrin specific peptidase 3 |
| chr14_-_32085595 | 0.01 |
ENSMUST00000022461.4
ENSMUST00000067955.5 ENSMUST00000124303.1 ENSMUST00000112000.1 |
Dph3
|
diphthamine biosynthesis 3 |
| chr9_+_122117258 | 0.01 |
ENSMUST00000146832.1
ENSMUST00000139181.1 |
Snrk
|
SNF related kinase |
| chr18_+_35771488 | 0.00 |
ENSMUST00000170693.1
|
Ube2d2a
|
ubiquitin-conjugating enzyme E2D 2A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gmeb2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 1.0 | 3.1 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 1.0 | 2.9 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.9 | 3.5 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.8 | 3.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.7 | 5.4 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.7 | 2.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.6 | 2.6 | GO:1904504 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.5 | 2.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.5 | 2.7 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.4 | 1.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.4 | 2.3 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.4 | 1.9 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.4 | 1.9 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.4 | 1.5 | GO:0071898 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.4 | 1.1 | GO:0034635 | glutathione transport(GO:0034635) oligopeptide transmembrane transport(GO:0035672) tripeptide transport(GO:0042939) |
| 0.3 | 1.3 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.3 | 0.3 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.3 | 1.1 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.2 | 1.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 1.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.2 | 1.0 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 0.7 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.2 | 1.6 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 2.0 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 0.6 | GO:0045041 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 1.0 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.2 | 1.2 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.2 | 0.6 | GO:2001074 | metanephric comma-shaped body morphogenesis(GO:0072278) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.2 | 2.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 0.8 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.2 | 1.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.9 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 2.7 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.1 | 2.8 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 | 2.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 1.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.9 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 1.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.3 | GO:1902445 | B cell negative selection(GO:0002352) B cell homeostatic proliferation(GO:0002358) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 2.7 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 1.0 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 1.3 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 | 1.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.9 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 1.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.4 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.5 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 0.7 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.2 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.3 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 1.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 1.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 2.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.8 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.9 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 1.9 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 3.9 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.9 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 1.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.8 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 2.4 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.3 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.6 | 1.9 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.6 | 3.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.6 | 2.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.6 | 2.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.5 | 2.7 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.4 | 1.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.4 | 1.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.3 | 1.6 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.3 | 2.7 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.2 | 3.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 2.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 0.8 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 1.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 2.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 1.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 2.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 1.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 3.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 2.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 4.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 3.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.9 | GO:0014704 | intercalated disc(GO:0014704) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0019202 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.7 | 2.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.6 | 2.6 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.6 | 2.6 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) bradykinin receptor binding(GO:0031711) |
| 0.6 | 1.9 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.4 | 1.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.4 | 2.7 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.4 | 2.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 1.9 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 1.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.3 | 1.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 3.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 1.0 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.2 | 0.7 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 4.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 2.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 1.5 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 0.8 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 1.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 1.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.4 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.1 | 2.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 1.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 1.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.6 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 3.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.7 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 2.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 2.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 5.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 3.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.4 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.6 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 7.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 5.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 5.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 5.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 3.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 1.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 4.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 5.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 2.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 3.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.3 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.6 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |