Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
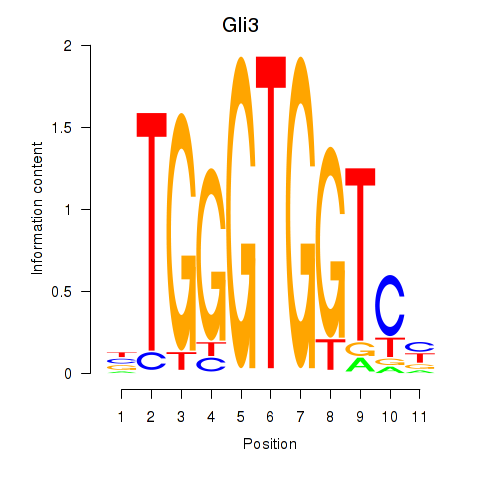
Results for Gli3_Zic1
Z-value: 1.52
Transcription factors associated with Gli3_Zic1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli3
|
ENSMUSG00000021318.9 | GLI-Kruppel family member GLI3 |
|
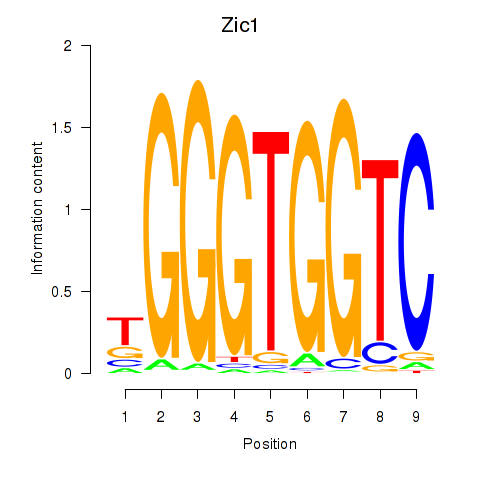
Zic1
|
ENSMUSG00000032368.8 | zinc finger protein of the cerebellum 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gli3 | mm10_v2_chr13_+_15463837_15463980 | 0.81 | 2.0e-09 | Click! |
| Zic1 | mm10_v2_chr9_-_91365778_91365815 | 0.49 | 2.5e-03 | Click! |
Activity profile of Gli3_Zic1 motif
Sorted Z-values of Gli3_Zic1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142657466 | 14.08 |
ENSMUST00000097936.2
ENSMUST00000000033.5 |
Igf2
|
insulin-like growth factor 2 |
| chr12_+_109453455 | 12.87 |
ENSMUST00000109844.4
ENSMUST00000109842.2 ENSMUST00000109843.1 ENSMUST00000109846.4 ENSMUST00000173539.1 ENSMUST00000109841.2 |
Dlk1
|
delta-like 1 homolog (Drosophila) |
| chr11_+_72999069 | 11.20 |
ENSMUST00000021141.7
|
P2rx1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr7_+_24370255 | 11.11 |
ENSMUST00000171904.1
|
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr6_+_30738044 | 9.34 |
ENSMUST00000128398.1
ENSMUST00000163949.2 ENSMUST00000124665.1 |
Mest
|
mesoderm specific transcript |
| chr11_-_102365111 | 9.10 |
ENSMUST00000006749.9
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr2_+_157560078 | 9.03 |
ENSMUST00000153739.2
ENSMUST00000173595.1 ENSMUST00000109526.1 ENSMUST00000173839.1 ENSMUST00000173041.1 ENSMUST00000173793.1 ENSMUST00000172487.1 ENSMUST00000088484.5 |
Nnat
|
neuronatin |
| chr6_+_113531675 | 8.81 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr9_-_103480328 | 7.53 |
ENSMUST00000124310.2
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr12_+_109540979 | 7.40 |
ENSMUST00000129245.1
ENSMUST00000143836.1 ENSMUST00000124106.1 |
Meg3
|
maternally expressed 3 |
| chr7_-_142656018 | 6.94 |
ENSMUST00000178921.1
|
Igf2
|
insulin-like growth factor 2 |
| chr12_+_109545390 | 6.65 |
ENSMUST00000146701.1
|
Meg3
|
maternally expressed 3 |
| chr13_+_44729535 | 6.43 |
ENSMUST00000174068.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr18_-_78142119 | 5.80 |
ENSMUST00000160639.1
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr7_-_24760311 | 5.47 |
ENSMUST00000063956.5
|
Cd177
|
CD177 antigen |
| chr8_+_23139064 | 5.41 |
ENSMUST00000033947.8
|
Ank1
|
ankyrin 1, erythroid |
| chr4_-_68954351 | 5.32 |
ENSMUST00000030036.5
|
Brinp1
|
bone morphogenic protein/retinoic acid inducible neural specific 1 |
| chr11_+_11684967 | 5.04 |
ENSMUST00000126058.1
ENSMUST00000141436.1 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr8_+_23139030 | 4.94 |
ENSMUST00000121075.1
|
Ank1
|
ankyrin 1, erythroid |
| chr10_-_127341583 | 4.42 |
ENSMUST00000026474.3
|
Gli1
|
GLI-Kruppel family member GLI1 |
| chrX_-_7574120 | 4.22 |
ENSMUST00000045924.7
ENSMUST00000115742.2 ENSMUST00000150787.1 |
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F |
| chr11_+_101176041 | 4.05 |
ENSMUST00000103109.3
|
Cntnap1
|
contactin associated protein-like 1 |
| chr1_-_133801031 | 4.05 |
ENSMUST00000143567.1
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr4_-_148130678 | 3.84 |
ENSMUST00000030862.4
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr7_+_99268338 | 3.68 |
ENSMUST00000107100.2
|
Map6
|
microtubule-associated protein 6 |
| chr11_+_72042455 | 3.67 |
ENSMUST00000021164.3
|
Fam64a
|
family with sequence similarity 64, member A |
| chr13_-_117025505 | 3.66 |
ENSMUST00000022239.6
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr17_+_25366550 | 3.66 |
ENSMUST00000069616.7
|
Tpsb2
|
tryptase beta 2 |
| chr18_-_35662180 | 3.65 |
ENSMUST00000025209.4
ENSMUST00000096573.2 |
Spata24
|
spermatogenesis associated 24 |
| chr10_-_79788924 | 3.62 |
ENSMUST00000020573.6
|
Prss57
|
protease, serine 57 |
| chr15_-_103310425 | 3.61 |
ENSMUST00000079824.4
|
Gpr84
|
G protein-coupled receptor 84 |
| chr15_-_100669496 | 3.56 |
ENSMUST00000182814.1
ENSMUST00000182068.1 |
Bin2
|
bridging integrator 2 |
| chr2_-_150668198 | 3.54 |
ENSMUST00000028944.3
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr15_-_100669512 | 3.51 |
ENSMUST00000182574.1
ENSMUST00000182775.1 |
Bin2
|
bridging integrator 2 |
| chr4_-_83021102 | 3.46 |
ENSMUST00000071708.5
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chrX_-_7964166 | 3.44 |
ENSMUST00000128449.1
|
Gata1
|
GATA binding protein 1 |
| chr8_-_71723308 | 3.42 |
ENSMUST00000125092.1
|
Fcho1
|
FCH domain only 1 |
| chr7_+_27499315 | 3.40 |
ENSMUST00000098644.2
ENSMUST00000108355.1 |
Prx
|
periaxin |
| chr3_-_69044697 | 3.34 |
ENSMUST00000136512.1
ENSMUST00000143454.1 ENSMUST00000107802.1 |
Trim59
|
tripartite motif-containing 59 |
| chr15_+_102296256 | 3.29 |
ENSMUST00000064924.4
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae) |
| chr13_+_108316395 | 3.21 |
ENSMUST00000171178.1
|
Depdc1b
|
DEP domain containing 1B |
| chr9_+_62858085 | 3.20 |
ENSMUST00000034777.6
ENSMUST00000163820.1 |
Calml4
|
calmodulin-like 4 |
| chr2_+_35282380 | 3.07 |
ENSMUST00000028239.6
|
Gsn
|
gelsolin |
| chrX_+_159303266 | 3.03 |
ENSMUST00000112491.1
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr2_+_118111876 | 3.02 |
ENSMUST00000039559.8
|
Thbs1
|
thrombospondin 1 |
| chr7_+_28540863 | 2.96 |
ENSMUST00000119180.2
|
Sycn
|
syncollin |
| chr15_+_99074968 | 2.95 |
ENSMUST00000039665.6
|
Troap
|
trophinin associated protein |
| chr4_-_4138432 | 2.90 |
ENSMUST00000070375.7
|
Penk
|
preproenkephalin |
| chr5_+_5573952 | 2.86 |
ENSMUST00000101627.2
|
Gm8773
|
predicted gene 8773 |
| chr9_-_123678782 | 2.86 |
ENSMUST00000170591.1
ENSMUST00000171647.1 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chrX_-_150812715 | 2.85 |
ENSMUST00000112697.3
|
Maged2
|
melanoma antigen, family D, 2 |
| chr9_-_123678873 | 2.75 |
ENSMUST00000040960.6
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr7_+_5056856 | 2.73 |
ENSMUST00000131368.1
ENSMUST00000123956.1 |
Ccdc106
|
coiled-coil domain containing 106 |
| chr15_-_95528702 | 2.64 |
ENSMUST00000166170.1
|
Nell2
|
NEL-like 2 |
| chr15_-_85581809 | 2.63 |
ENSMUST00000023015.7
|
Wnt7b
|
wingless-related MMTV integration site 7B |
| chr1_-_75505641 | 2.63 |
ENSMUST00000155084.1
|
Obsl1
|
obscurin-like 1 |
| chr17_+_56304313 | 2.63 |
ENSMUST00000113035.1
ENSMUST00000113039.2 ENSMUST00000142387.1 |
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr11_+_69095217 | 2.50 |
ENSMUST00000101004.2
|
Per1
|
period circadian clock 1 |
| chr2_-_156839790 | 2.48 |
ENSMUST00000134838.1
ENSMUST00000137463.1 ENSMUST00000149275.2 |
Gm14230
|
predicted gene 14230 |
| chr4_-_117178726 | 2.48 |
ENSMUST00000153953.1
ENSMUST00000106436.1 |
Kif2c
|
kinesin family member 2C |
| chr11_-_83462961 | 2.47 |
ENSMUST00000021020.6
ENSMUST00000119346.1 ENSMUST00000103209.3 ENSMUST00000108137.2 |
Mmp28
|
matrix metallopeptidase 28 (epilysin) |
| chr4_+_44943727 | 2.45 |
ENSMUST00000154177.1
|
Gm12678
|
predicted gene 12678 |
| chr5_-_24730635 | 2.43 |
ENSMUST00000068693.5
|
Wdr86
|
WD repeat domain 86 |
| chr1_+_129273344 | 2.41 |
ENSMUST00000073527.6
ENSMUST00000040311.7 |
Thsd7b
|
thrombospondin, type I, domain containing 7B |
| chr9_-_35558522 | 2.36 |
ENSMUST00000034612.5
|
Ddx25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25 |
| chr4_+_43957401 | 2.35 |
ENSMUST00000030202.7
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr9_-_108190352 | 2.33 |
ENSMUST00000035208.7
|
Bsn
|
bassoon |
| chr7_-_126704816 | 2.31 |
ENSMUST00000032949.7
|
Coro1a
|
coronin, actin binding protein 1A |
| chr2_-_91931675 | 2.30 |
ENSMUST00000111309.1
|
Mdk
|
midkine |
| chr8_+_123411424 | 2.26 |
ENSMUST00000071134.3
|
Tubb3
|
tubulin, beta 3 class III |
| chr1_-_75506331 | 2.23 |
ENSMUST00000113567.2
ENSMUST00000113565.2 |
Obsl1
|
obscurin-like 1 |
| chr7_-_44227421 | 2.20 |
ENSMUST00000072123.4
|
Gm10109
|
predicted gene 10109 |
| chr2_-_92371039 | 2.19 |
ENSMUST00000068586.6
|
Gyltl1b
|
glycosyltransferase-like 1B |
| chr10_+_79777261 | 2.18 |
ENSMUST00000020575.4
|
Fstl3
|
follistatin-like 3 |
| chr11_+_75656103 | 2.16 |
ENSMUST00000136935.1
|
Myo1c
|
myosin IC |
| chr11_+_96282648 | 2.16 |
ENSMUST00000168043.1
|
Hoxb8
|
homeobox B8 |
| chr8_-_105637403 | 2.16 |
ENSMUST00000182046.1
|
Gm5914
|
predicted gene 5914 |
| chr12_+_103388656 | 2.15 |
ENSMUST00000101094.2
ENSMUST00000021620.6 |
Otub2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr13_-_99516537 | 2.15 |
ENSMUST00000064762.4
|
Map1b
|
microtubule-associated protein 1B |
| chr3_+_31149934 | 2.13 |
ENSMUST00000046174.6
|
Cldn11
|
claudin 11 |
| chrX_+_74329058 | 2.12 |
ENSMUST00000004326.3
|
Plxna3
|
plexin A3 |
| chr11_-_78245942 | 2.08 |
ENSMUST00000002121.4
|
Supt6
|
suppressor of Ty 6 |
| chr7_-_126704736 | 2.06 |
ENSMUST00000131415.1
|
Coro1a
|
coronin, actin binding protein 1A |
| chr17_-_25727364 | 2.01 |
ENSMUST00000170070.1
ENSMUST00000048054.7 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr7_+_27258725 | 2.01 |
ENSMUST00000079258.6
|
Numbl
|
numb-like |
| chr12_-_32953772 | 1.99 |
ENSMUST00000180391.1
ENSMUST00000181670.1 |
4933406C10Rik
|
RIKEN cDNA 4933406C10 gene |
| chr11_+_70000578 | 1.95 |
ENSMUST00000019362.8
|
Dvl2
|
dishevelled 2, dsh homolog (Drosophila) |
| chr11_+_100415722 | 1.94 |
ENSMUST00000107400.2
|
Fkbp10
|
FK506 binding protein 10 |
| chrX_-_150812932 | 1.93 |
ENSMUST00000131241.1
ENSMUST00000147152.1 ENSMUST00000143843.1 |
Maged2
|
melanoma antigen, family D, 2 |
| chr7_-_118443549 | 1.91 |
ENSMUST00000081574.4
|
Syt17
|
synaptotagmin XVII |
| chr5_-_117319242 | 1.91 |
ENSMUST00000100834.1
|
Gm10399
|
predicted gene 10399 |
| chr7_+_5056706 | 1.90 |
ENSMUST00000144802.1
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr2_+_29889720 | 1.85 |
ENSMUST00000113767.1
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr18_-_21652362 | 1.82 |
ENSMUST00000049105.4
|
Klhl14
|
kelch-like 14 |
| chr6_-_125494754 | 1.80 |
ENSMUST00000032492.8
|
Cd9
|
CD9 antigen |
| chr15_+_78899755 | 1.80 |
ENSMUST00000001226.3
ENSMUST00000061239.7 ENSMUST00000109698.2 |
Sh3bp1
|
SH3-domain binding protein 1 |
| chr8_+_68880491 | 1.79 |
ENSMUST00000015712.8
|
Lpl
|
lipoprotein lipase |
| chr11_+_115154139 | 1.79 |
ENSMUST00000021076.5
|
Rab37
|
RAB37, member of RAS oncogene family |
| chr15_-_95528228 | 1.78 |
ENSMUST00000075275.2
|
Nell2
|
NEL-like 2 |
| chr6_-_118780324 | 1.76 |
ENSMUST00000112793.3
ENSMUST00000075591.6 ENSMUST00000078320.7 ENSMUST00000112790.2 |
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr4_+_43957678 | 1.76 |
ENSMUST00000107855.1
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr8_+_70501116 | 1.76 |
ENSMUST00000127983.1
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr3_+_105904377 | 1.71 |
ENSMUST00000000574.1
|
Adora3
|
adenosine A3 receptor |
| chr2_+_29890096 | 1.71 |
ENSMUST00000113762.1
ENSMUST00000113765.1 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr1_+_65311257 | 1.70 |
ENSMUST00000027083.6
|
Pth2r
|
parathyroid hormone 2 receptor |
| chr4_+_154869585 | 1.68 |
ENSMUST00000079269.7
ENSMUST00000163732.1 ENSMUST00000080559.6 |
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr7_+_99535439 | 1.65 |
ENSMUST00000098266.2
ENSMUST00000179755.1 |
Arrb1
|
arrestin, beta 1 |
| chr2_+_29889785 | 1.64 |
ENSMUST00000113763.1
ENSMUST00000113757.1 ENSMUST00000113756.1 ENSMUST00000133233.1 ENSMUST00000113759.2 ENSMUST00000113755.1 ENSMUST00000137558.1 ENSMUST00000046571.7 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr11_-_45955465 | 1.64 |
ENSMUST00000011398.6
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr10_+_80016653 | 1.63 |
ENSMUST00000099501.3
|
Hmha1
|
histocompatibility (minor) HA-1 |
| chr3_-_92485886 | 1.63 |
ENSMUST00000054599.7
|
Sprr1a
|
small proline-rich protein 1A |
| chr8_-_105637350 | 1.61 |
ENSMUST00000182863.1
|
Gm5914
|
predicted gene 5914 |
| chr11_+_96282529 | 1.59 |
ENSMUST00000125410.1
|
Hoxb8
|
homeobox B8 |
| chr13_+_44729794 | 1.58 |
ENSMUST00000172830.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr10_+_80016901 | 1.57 |
ENSMUST00000105373.1
|
Hmha1
|
histocompatibility (minor) HA-1 |
| chr9_-_54661870 | 1.57 |
ENSMUST00000034822.5
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr11_-_34833631 | 1.56 |
ENSMUST00000093191.2
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr7_+_125603420 | 1.54 |
ENSMUST00000033000.6
|
Il21r
|
interleukin 21 receptor |
| chr14_-_54926784 | 1.54 |
ENSMUST00000022813.6
|
Efs
|
embryonal Fyn-associated substrate |
| chr2_+_156840077 | 1.53 |
ENSMUST00000081335.6
ENSMUST00000073352.3 |
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr1_+_167001457 | 1.53 |
ENSMUST00000126198.1
|
Fam78b
|
family with sequence similarity 78, member B |
| chr13_-_49320219 | 1.52 |
ENSMUST00000110086.1
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr15_-_100669535 | 1.49 |
ENSMUST00000183211.1
|
Bin2
|
bridging integrator 2 |
| chr2_+_29890534 | 1.48 |
ENSMUST00000113764.3
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr11_+_87760533 | 1.46 |
ENSMUST00000039627.5
ENSMUST00000100644.3 |
Bzrap1
|
benzodiazepine receptor associated protein 1 |
| chr4_-_136956784 | 1.45 |
ENSMUST00000030420.8
|
Epha8
|
Eph receptor A8 |
| chr2_+_29890063 | 1.44 |
ENSMUST00000028128.6
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr7_-_25297866 | 1.42 |
ENSMUST00000148150.1
ENSMUST00000155118.1 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr8_+_120002720 | 1.41 |
ENSMUST00000108972.3
|
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr11_-_61494173 | 1.40 |
ENSMUST00000101085.2
ENSMUST00000079080.6 ENSMUST00000108714.1 |
Mapk7
|
mitogen-activated protein kinase 7 |
| chr12_-_4233958 | 1.40 |
ENSMUST00000111169.3
ENSMUST00000020981.5 |
Cenpo
|
centromere protein O |
| chr14_-_33185066 | 1.37 |
ENSMUST00000061753.8
ENSMUST00000130509.2 |
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr5_+_43818893 | 1.36 |
ENSMUST00000101237.4
|
Bst1
|
bone marrow stromal cell antigen 1 |
| chr11_-_76399107 | 1.36 |
ENSMUST00000021204.3
|
Nxn
|
nucleoredoxin |
| chr2_+_105127200 | 1.36 |
ENSMUST00000139585.1
|
Wt1
|
Wilms tumor 1 homolog |
| chr17_+_34039437 | 1.33 |
ENSMUST00000131134.1
ENSMUST00000087497.4 ENSMUST00000114255.1 ENSMUST00000114252.1 |
Col11a2
|
collagen, type XI, alpha 2 |
| chrX_+_159697308 | 1.32 |
ENSMUST00000123433.1
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr13_+_6548154 | 1.32 |
ENSMUST00000021611.8
|
Pitrm1
|
pitrilysin metallepetidase 1 |
| chr18_+_62180119 | 1.31 |
ENSMUST00000067743.1
|
Gm9949
|
predicted gene 9949 |
| chr7_+_80186835 | 1.30 |
ENSMUST00000107383.1
ENSMUST00000032754.7 |
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr5_-_115300912 | 1.30 |
ENSMUST00000112090.1
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr14_+_84443553 | 1.30 |
ENSMUST00000071370.5
|
Pcdh17
|
protocadherin 17 |
| chr17_-_15041457 | 1.29 |
ENSMUST00000097400.4
ENSMUST00000097398.4 ENSMUST00000040746.6 |
Gm3448
Tcte3
|
predicted gene 3448 t-complex-associated testis expressed 3 |
| chr5_-_5266038 | 1.28 |
ENSMUST00000115451.1
ENSMUST00000115452.1 ENSMUST00000131392.1 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr11_+_53519725 | 1.28 |
ENSMUST00000108987.1
ENSMUST00000121334.1 ENSMUST00000117061.1 |
Sept8
|
septin 8 |
| chr5_-_115300957 | 1.28 |
ENSMUST00000009157.3
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr1_+_72824482 | 1.27 |
ENSMUST00000047328.4
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr1_-_133906973 | 1.26 |
ENSMUST00000126123.1
|
Optc
|
opticin |
| chr7_+_25267669 | 1.23 |
ENSMUST00000169266.1
|
Cic
|
capicua homolog (Drosophila) |
| chr6_+_125067913 | 1.22 |
ENSMUST00000088292.5
|
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr2_-_153444441 | 1.22 |
ENSMUST00000109784.1
|
8430427H17Rik
|
RIKEN cDNA 8430427H17 gene |
| chr13_+_44730726 | 1.22 |
ENSMUST00000173704.1
ENSMUST00000044608.7 ENSMUST00000173367.1 |
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr11_+_77982710 | 1.22 |
ENSMUST00000108360.1
ENSMUST00000049167.7 |
Phf12
|
PHD finger protein 12 |
| chr3_+_92288566 | 1.22 |
ENSMUST00000090872.4
|
Sprr2a3
|
small proline-rich protein 2A3 |
| chr6_+_29694204 | 1.21 |
ENSMUST00000046750.7
ENSMUST00000115250.3 |
Tspan33
|
tetraspanin 33 |
| chr2_-_113848655 | 1.21 |
ENSMUST00000102545.1
ENSMUST00000110948.1 |
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr17_-_44735612 | 1.20 |
ENSMUST00000162373.1
ENSMUST00000162878.1 |
Runx2
|
runt related transcription factor 2 |
| chr9_-_44234014 | 1.20 |
ENSMUST00000037644.6
|
Cbl
|
Casitas B-lineage lymphoma |
| chrX_+_140956892 | 1.20 |
ENSMUST00000112971.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chrX_-_94542037 | 1.19 |
ENSMUST00000026142.7
|
Maged1
|
melanoma antigen, family D, 1 |
| chr15_-_56694525 | 1.17 |
ENSMUST00000050544.7
|
Has2
|
hyaluronan synthase 2 |
| chr11_-_106349389 | 1.15 |
ENSMUST00000021056.7
|
Scn4a
|
sodium channel, voltage-gated, type IV, alpha |
| chr2_+_174643534 | 1.14 |
ENSMUST00000059452.5
|
Zfp831
|
zinc finger protein 831 |
| chr7_-_25297967 | 1.13 |
ENSMUST00000005583.5
|
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr11_+_115462464 | 1.12 |
ENSMUST00000106532.3
ENSMUST00000092445.5 ENSMUST00000153466.1 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr1_+_43445736 | 1.11 |
ENSMUST00000086421.5
ENSMUST00000114744.1 |
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2 |
| chr8_-_22125030 | 1.11 |
ENSMUST00000169834.1
|
Nek5
|
NIMA (never in mitosis gene a)-related expressed kinase 5 |
| chr2_-_113848601 | 1.10 |
ENSMUST00000110949.2
|
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr13_-_23551648 | 1.10 |
ENSMUST00000102971.1
|
Hist1h4f
|
histone cluster 1, H4f |
| chrX_-_53114530 | 1.10 |
ENSMUST00000114843.2
|
Plac1
|
placental specific protein 1 |
| chr18_-_62179948 | 1.10 |
ENSMUST00000053640.3
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr7_+_130865835 | 1.10 |
ENSMUST00000075181.4
ENSMUST00000048180.5 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr9_-_50752348 | 1.09 |
ENSMUST00000042790.3
|
Hspb2
|
heat shock protein 2 |
| chr8_+_121127827 | 1.08 |
ENSMUST00000181609.1
|
Foxl1
|
forkhead box L1 |
| chr6_-_28830345 | 1.08 |
ENSMUST00000171353.1
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr2_+_106693185 | 1.07 |
ENSMUST00000111063.1
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr7_+_128129536 | 1.07 |
ENSMUST00000033053.6
|
Itgax
|
integrin alpha X |
| chr11_-_70229677 | 1.07 |
ENSMUST00000153449.1
ENSMUST00000000326.5 |
Bcl6b
|
B cell CLL/lymphoma 6, member B |
| chr6_-_101377342 | 1.07 |
ENSMUST00000151175.1
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr15_+_40655020 | 1.06 |
ENSMUST00000053467.4
|
Zfpm2
|
zinc finger protein, multitype 2 |
| chr6_-_148944750 | 1.05 |
ENSMUST00000111562.1
ENSMUST00000081956.5 |
Fam60a
|
family with sequence similarity 60, member A |
| chr11_+_73018986 | 1.05 |
ENSMUST00000092937.6
|
Camkk1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr5_-_38561658 | 1.04 |
ENSMUST00000005234.9
|
Wdr1
|
WD repeat domain 1 |
| chr5_+_117319292 | 1.03 |
ENSMUST00000086464.4
|
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr8_+_83608175 | 1.03 |
ENSMUST00000005620.8
|
Dnajb1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr8_+_23139157 | 1.01 |
ENSMUST00000174435.1
|
Ank1
|
ankyrin 1, erythroid |
| chr17_+_34894515 | 1.00 |
ENSMUST00000052778.8
|
Zbtb12
|
zinc finger and BTB domain containing 12 |
| chr6_-_52218686 | 1.00 |
ENSMUST00000134367.2
|
Hoxa7
|
homeobox A7 |
| chr3_-_116253467 | 1.00 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr4_+_123233556 | 1.00 |
ENSMUST00000040821.4
|
Heyl
|
hairy/enhancer-of-split related with YRPW motif-like |
| chr11_+_49901803 | 0.99 |
ENSMUST00000093141.5
ENSMUST00000093142.5 |
Rasgef1c
|
RasGEF domain family, member 1C |
| chr18_+_52767994 | 0.99 |
ENSMUST00000025413.7
ENSMUST00000163742.2 ENSMUST00000178011.1 |
Sncaip
|
synuclein, alpha interacting protein (synphilin) |
| chr19_+_8802486 | 0.99 |
ENSMUST00000172175.1
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr2_-_59948155 | 0.97 |
ENSMUST00000153136.1
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr7_+_128246953 | 0.96 |
ENSMUST00000167965.1
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr12_+_32378692 | 0.96 |
ENSMUST00000172332.2
|
Ccdc71l
|
coiled-coil domain containing 71 like |
| chr19_+_6384395 | 0.96 |
ENSMUST00000035269.8
ENSMUST00000113483.1 |
Pygm
|
muscle glycogen phosphorylase |
| chr2_-_33431324 | 0.96 |
ENSMUST00000113158.1
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr6_-_41636389 | 0.96 |
ENSMUST00000031902.5
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr14_+_25607797 | 0.96 |
ENSMUST00000160229.1
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gli3_Zic1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 21.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 2.4 | 7.1 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 2.2 | 11.2 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 1.9 | 5.8 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 1.7 | 5.0 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 1.4 | 5.6 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 1.1 | 3.4 | GO:0030221 | basophil differentiation(GO:0030221) |
| 1.1 | 4.4 | GO:0060032 | notochord regression(GO:0060032) |
| 1.0 | 3.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of establishment of T cell polarity(GO:1903903) |
| 1.0 | 5.9 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 1.0 | 9.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.9 | 11.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.9 | 3.4 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.8 | 0.8 | GO:0060915 | fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.8 | 2.3 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.7 | 4.4 | GO:0032796 | uropod organization(GO:0032796) |
| 0.7 | 2.0 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.7 | 2.6 | GO:0072054 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) renal outer medulla development(GO:0072054) inner medullary collecting duct development(GO:0072061) |
| 0.6 | 1.9 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.6 | 9.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.6 | 4.2 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.6 | 4.8 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.6 | 3.5 | GO:0051790 | acetate metabolic process(GO:0006083) short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.5 | 3.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.5 | 13.5 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.5 | 2.5 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.5 | 2.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.5 | 2.5 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.5 | 2.9 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.5 | 8.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.5 | 14.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.5 | 1.4 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.5 | 1.4 | GO:2001076 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.4 | 1.3 | GO:1904978 | regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.4 | 2.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.4 | 1.2 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.4 | 3.0 | GO:1904378 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.4 | 1.8 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.4 | 2.1 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.4 | 1.8 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.3 | 9.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 7.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.3 | 2.3 | GO:0030421 | defecation(GO:0030421) |
| 0.3 | 0.3 | GO:1904907 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.3 | 4.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 2.8 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 0.9 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.3 | 0.9 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.3 | 8.7 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.3 | 1.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.3 | 4.4 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.3 | 1.7 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.3 | 1.1 | GO:1904504 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.3 | 1.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 | 1.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 1.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.3 | 1.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.3 | 11.4 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.3 | 1.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 1.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 1.0 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.2 | 1.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 2.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.2 | 2.6 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.9 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.2 | 1.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 3.3 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 0.7 | GO:0071288 | cellular response to mercury ion(GO:0071288) cellular response to water stimulus(GO:0071462) |
| 0.2 | 1.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 4.3 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 0.6 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.2 | 0.2 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.2 | 1.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.2 | 0.5 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.2 | 2.9 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.2 | 1.7 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.2 | 0.8 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.2 | 0.3 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.2 | 1.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 2.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 0.8 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.2 | 0.8 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.2 | 0.3 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.2 | 7.8 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.2 | 2.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 3.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.4 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 1.3 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 1.5 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 3.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.6 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 2.6 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 1.0 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.9 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.5 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 2.1 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.1 | 0.8 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 3.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.5 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.1 | 0.1 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.1 | 1.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 1.1 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 | 1.8 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 8.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.7 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.4 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.7 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 1.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.7 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 0.3 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 1.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.2 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 1.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.2 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.1 | 0.5 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.0 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 1.1 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 1.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.2 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.1 | 0.3 | GO:0010841 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 1.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.3 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 3.0 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 2.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.1 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.8 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 2.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.3 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.1 | 1.3 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.1 | 0.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 3.8 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.2 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 | 1.2 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.5 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 0.7 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 0.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 4.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.4 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.5 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.4 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 1.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.2 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 1.0 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.4 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 1.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 2.3 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.4 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 0.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.4 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 1.5 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 1.1 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.1 | GO:1905034 | regulation of defense response to fungus(GO:1900150) regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.0 | 0.6 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 3.2 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 | 0.5 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 2.4 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.1 | GO:0097240 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.6 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 1.2 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 1.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.6 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.3 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 1.0 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.5 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.7 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.5 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.5 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.2 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 1.2 | 3.6 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.8 | 2.3 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.8 | 3.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.7 | 2.9 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.5 | 8.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.5 | 12.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.5 | 1.8 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.4 | 12.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 0.9 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.4 | 3.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.3 | 3.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 9.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 4.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 4.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.3 | 0.9 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.3 | 2.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 1.0 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.3 | 1.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 4.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 2.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 1.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.2 | 2.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 4.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 1.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 3.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.9 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 2.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 4.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.7 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 1.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 1.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 9.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.8 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 2.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 1.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 2.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 3.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 5.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 2.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 3.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 7.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 3.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 15.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.9 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 5.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 2.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 7.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 3.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 3.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 1.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 1.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.6 | 11.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 1.2 | 3.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.1 | 10.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.1 | 4.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 1.0 | 3.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.0 | 5.8 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.9 | 2.6 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.8 | 2.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.8 | 3.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.7 | 3.7 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.7 | 2.6 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.6 | 23.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.6 | 4.0 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.5 | 5.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.4 | 8.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 2.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 6.6 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.4 | 9.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.4 | 8.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.4 | 2.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 1.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 1.4 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.3 | 1.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.3 | 1.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 0.9 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.3 | 1.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 1.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 1.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) bradykinin receptor binding(GO:0031711) |
| 0.3 | 2.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 1.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.3 | 1.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.3 | 4.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 1.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.6 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 2.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.2 | 1.8 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 0.7 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.2 | 3.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 4.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 9.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.2 | 1.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 2.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 3.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 5.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.2 | 3.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 1.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 4.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 1.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 2.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 2.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 2.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 12.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.3 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.7 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.1 | 0.8 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.9 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 1.9 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 1.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 1.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.6 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.1 | 0.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 1.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.4 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.7 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 3.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.4 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 9.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 4.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 1.1 | GO:1905030 | voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 9.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 3.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 1.9 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 1.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 17.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 6.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.9 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 2.5 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 11.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 12.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 8.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 4.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 4.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 3.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 6.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 1.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 3.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 5.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 10.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 1.1 | 22.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.7 | 8.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.6 | 3.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 14.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 9.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 3.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 4.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 4.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 3.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 10.3 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 1.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 4.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 3.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 11.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 5.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 4.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 4.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.8 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 2.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |