Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
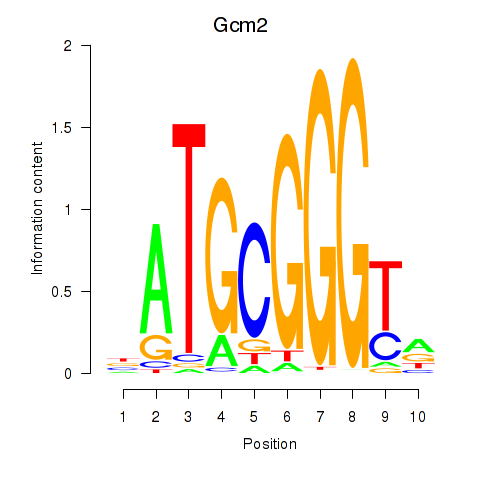
Results for Gcm2
Z-value: 1.00
Transcription factors associated with Gcm2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gcm2
|
ENSMUSG00000021362.6 | glial cells missing homolog 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gcm2 | mm10_v2_chr13_-_41109988_41109988 | 0.25 | 1.5e-01 | Click! |
Activity profile of Gcm2 motif
Sorted Z-values of Gcm2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_111691002 | 7.01 |
ENSMUST00000034435.5
|
Ctrb1
|
chymotrypsinogen B1 |
| chr3_-_113258837 | 4.03 |
ENSMUST00000098673.3
|
Amy2a5
|
amylase 2a5 |
| chr11_-_87875524 | 3.41 |
ENSMUST00000049768.3
|
Epx
|
eosinophil peroxidase |
| chr4_-_141825997 | 2.88 |
ENSMUST00000102481.3
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr6_+_78425973 | 2.84 |
ENSMUST00000079926.5
|
Reg1
|
regenerating islet-derived 1 |
| chr6_-_41314700 | 2.61 |
ENSMUST00000064324.5
|
Try5
|
trypsin 5 |
| chr6_+_41302265 | 2.18 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr1_+_93006328 | 2.10 |
ENSMUST00000059676.4
|
Aqp12
|
aquaporin 12 |
| chr7_-_142657466 | 2.03 |
ENSMUST00000097936.2
ENSMUST00000000033.5 |
Igf2
|
insulin-like growth factor 2 |
| chr7_-_142578139 | 1.99 |
ENSMUST00000136359.1
|
H19
|
H19 fetal liver mRNA |
| chr19_-_39886730 | 1.94 |
ENSMUST00000168838.1
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr3_-_113324052 | 1.87 |
ENSMUST00000179314.1
|
Amy2a3
|
amylase 2a3 |
| chr19_+_54045182 | 1.79 |
ENSMUST00000036700.5
|
Adra2a
|
adrenergic receptor, alpha 2a |
| chr7_-_142656018 | 1.65 |
ENSMUST00000178921.1
|
Igf2
|
insulin-like growth factor 2 |
| chr7_-_13989588 | 1.63 |
ENSMUST00000165167.1
ENSMUST00000108520.2 |
Sult2a4
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 4 |
| chr12_+_110279228 | 1.63 |
ENSMUST00000097228.4
|
Dio3
|
deiodinase, iodothyronine type III |
| chr15_-_44788016 | 1.59 |
ENSMUST00000090057.4
ENSMUST00000110269.1 |
Sybu
|
syntabulin (syntaxin-interacting) |
| chr16_+_22951072 | 1.47 |
ENSMUST00000023590.8
|
Hrg
|
histidine-rich glycoprotein |
| chr2_-_164745916 | 1.41 |
ENSMUST00000109328.1
ENSMUST00000043448.1 |
Wfdc3
Wfdc3
|
WAP four-disulfide core domain 3 WAP four-disulfide core domain 3 |
| chr9_+_90162978 | 1.34 |
ENSMUST00000113060.1
|
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr9_+_90163057 | 1.32 |
ENSMUST00000113059.1
ENSMUST00000167122.1 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr9_+_111019284 | 1.32 |
ENSMUST00000035077.3
|
Ltf
|
lactotransferrin |
| chr12_-_79007276 | 1.29 |
ENSMUST00000056660.6
ENSMUST00000174721.1 |
Tmem229b
|
transmembrane protein 229B |
| chr15_+_44787746 | 1.26 |
ENSMUST00000181839.1
|
2310069G16Rik
|
RIKEN cDNA 2310069G16 gene |
| chr14_-_60177482 | 1.26 |
ENSMUST00000140924.1
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr1_-_184999549 | 1.25 |
ENSMUST00000027929.4
|
Mark1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr5_+_141241490 | 1.24 |
ENSMUST00000085774.4
|
Sdk1
|
sidekick homolog 1 (chicken) |
| chr13_-_62888282 | 1.23 |
ENSMUST00000092888.4
|
Fbp1
|
fructose bisphosphatase 1 |
| chr17_+_24696234 | 1.22 |
ENSMUST00000019464.7
|
Noxo1
|
NADPH oxidase organizer 1 |
| chr10_+_95417352 | 1.18 |
ENSMUST00000181781.1
|
5730420D15Rik
|
RIKEN cDNA 5730420D15 gene |
| chr19_-_4059295 | 1.17 |
ENSMUST00000076451.6
|
BC021614
|
cDNA sequence BC021614 |
| chr9_+_45138437 | 1.13 |
ENSMUST00000060125.5
|
Scn4b
|
sodium channel, type IV, beta |
| chr15_-_89410230 | 1.13 |
ENSMUST00000109314.2
|
Syce3
|
synaptonemal complex central element protein 3 |
| chr7_-_143460989 | 1.12 |
ENSMUST00000167912.1
ENSMUST00000037287.6 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C (P57) |
| chr17_-_43502773 | 1.09 |
ENSMUST00000024707.8
ENSMUST00000117137.1 |
Mep1a
|
meprin 1 alpha |
| chr12_-_103989950 | 1.06 |
ENSMUST00000120251.2
|
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr12_-_115964196 | 1.05 |
ENSMUST00000103550.2
|
Ighv1-83
|
immunoglobulin heavy variable 1-83 |
| chrX_-_20291776 | 1.03 |
ENSMUST00000072451.4
|
Slc9a7
|
solute carrier family 9 (sodium/hydrogen exchanger), member 7 |
| chr7_+_45705088 | 1.03 |
ENSMUST00000080885.3
|
Dbp
|
D site albumin promoter binding protein |
| chr9_+_46268601 | 1.00 |
ENSMUST00000121598.1
|
Apoa5
|
apolipoprotein A-V |
| chr9_-_65391652 | 0.97 |
ENSMUST00000068307.3
|
Kbtbd13
|
kelch repeat and BTB (POZ) domain containing 13 |
| chr5_+_100846364 | 0.97 |
ENSMUST00000092990.3
ENSMUST00000145612.1 |
Agpat9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr9_+_36832684 | 0.97 |
ENSMUST00000034630.8
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr6_-_85374606 | 0.96 |
ENSMUST00000060837.7
|
Rab11fip5
|
RAB11 family interacting protein 5 (class I) |
| chr4_-_32602760 | 0.95 |
ENSMUST00000056517.2
|
Gja10
|
gap junction protein, alpha 10 |
| chr17_-_7352696 | 0.94 |
ENSMUST00000115747.1
|
Ttll2
|
tubulin tyrosine ligase-like family, member 2 |
| chr12_-_76709997 | 0.94 |
ENSMUST00000166101.1
|
Sptb
|
spectrin beta, erythrocytic |
| chr12_-_103242143 | 0.93 |
ENSMUST00000074416.3
|
Prima1
|
proline rich membrane anchor 1 |
| chr12_+_9574437 | 0.92 |
ENSMUST00000057021.7
|
Osr1
|
odd-skipped related 1 (Drosophila) |
| chr11_+_83709015 | 0.90 |
ENSMUST00000001009.7
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr1_+_134182150 | 0.90 |
ENSMUST00000156873.1
|
Chi3l1
|
chitinase 3-like 1 |
| chr12_-_103989917 | 0.90 |
ENSMUST00000151709.2
ENSMUST00000176246.1 ENSMUST00000074693.7 |
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr18_-_13972617 | 0.89 |
ENSMUST00000025288.7
|
Zfp521
|
zinc finger protein 521 |
| chr7_-_38107490 | 0.86 |
ENSMUST00000108023.3
|
Ccne1
|
cyclin E1 |
| chr5_+_90561102 | 0.86 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr6_+_86628174 | 0.84 |
ENSMUST00000043400.6
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr19_-_10604258 | 0.82 |
ENSMUST00000037678.6
|
Dak
|
dihydroxyacetone kinase 2 homolog (yeast) |
| chrX_-_52613936 | 0.82 |
ENSMUST00000114857.1
|
Gpc3
|
glypican 3 |
| chr6_-_29179584 | 0.81 |
ENSMUST00000159200.1
|
Prrt4
|
proline-rich transmembrane protein 4 |
| chr9_-_35558522 | 0.81 |
ENSMUST00000034612.5
|
Ddx25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25 |
| chr5_+_118027743 | 0.80 |
ENSMUST00000031304.7
|
Tesc
|
tescalcin |
| chr7_-_105600103 | 0.80 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr14_+_118787894 | 0.80 |
ENSMUST00000047761.6
ENSMUST00000071546.7 |
Cldn10
|
claudin 10 |
| chr14_+_70555900 | 0.80 |
ENSMUST00000163060.1
|
Hr
|
hairless |
| chrX_-_52613913 | 0.79 |
ENSMUST00000069360.7
|
Gpc3
|
glypican 3 |
| chr5_+_100846130 | 0.79 |
ENSMUST00000112887.1
ENSMUST00000031255.8 |
Agpat9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr11_+_104550663 | 0.78 |
ENSMUST00000018800.2
|
Myl4
|
myosin, light polypeptide 4 |
| chr15_-_74734313 | 0.77 |
ENSMUST00000023260.3
|
Lypd2
|
Ly6/Plaur domain containing 2 |
| chr15_-_99457712 | 0.76 |
ENSMUST00000161948.1
|
Nckap5l
|
NCK-associated protein 5-like |
| chr11_-_83530505 | 0.76 |
ENSMUST00000035938.2
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr17_-_84682932 | 0.76 |
ENSMUST00000066175.3
|
Abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr1_+_132298606 | 0.75 |
ENSMUST00000046071.4
|
Klhdc8a
|
kelch domain containing 8A |
| chr15_-_86186136 | 0.75 |
ENSMUST00000044332.9
|
Cerk
|
ceramide kinase |
| chr9_+_72925622 | 0.74 |
ENSMUST00000038489.5
|
Pygo1
|
pygopus 1 |
| chr5_+_33983534 | 0.73 |
ENSMUST00000114382.1
|
Gm1673
|
predicted gene 1673 |
| chr7_-_105787544 | 0.73 |
ENSMUST00000078482.5
ENSMUST00000154659.1 |
Dchs1
|
dachsous 1 (Drosophila) |
| chr9_+_75625707 | 0.73 |
ENSMUST00000034702.4
|
Lysmd2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr4_+_141239499 | 0.72 |
ENSMUST00000141834.2
|
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr3_+_145292472 | 0.72 |
ENSMUST00000029848.4
ENSMUST00000139001.1 |
Col24a1
|
collagen, type XXIV, alpha 1 |
| chr2_+_25289899 | 0.72 |
ENSMUST00000028337.6
|
Lrrc26
|
leucine rich repeat containing 26 |
| chr17_+_30901811 | 0.71 |
ENSMUST00000114574.1
|
Glp1r
|
glucagon-like peptide 1 receptor |
| chr8_-_25038875 | 0.71 |
ENSMUST00000084031.4
|
Htra4
|
HtrA serine peptidase 4 |
| chr2_+_147364989 | 0.70 |
ENSMUST00000109968.2
|
Pax1
|
paired box gene 1 |
| chr2_-_32431104 | 0.70 |
ENSMUST00000127961.1
ENSMUST00000136361.1 ENSMUST00000052119.7 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr5_+_136693146 | 0.70 |
ENSMUST00000005611.5
|
Myl10
|
myosin, light chain 10, regulatory |
| chr16_+_30008657 | 0.70 |
ENSMUST00000181485.1
|
4632428C04Rik
|
RIKEN cDNA 4632428C04 gene |
| chr15_+_99579054 | 0.69 |
ENSMUST00000023752.4
|
Aqp2
|
aquaporin 2 |
| chr14_-_56262233 | 0.69 |
ENSMUST00000015581.4
|
Gzmb
|
granzyme B |
| chr9_+_57940104 | 0.69 |
ENSMUST00000043059.7
|
Sema7a
|
sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A |
| chr2_+_156840966 | 0.68 |
ENSMUST00000109564.1
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr7_+_30291659 | 0.68 |
ENSMUST00000014065.8
ENSMUST00000150892.1 ENSMUST00000126216.1 |
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr8_+_23411490 | 0.68 |
ENSMUST00000033952.7
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr10_+_21690845 | 0.68 |
ENSMUST00000071008.3
|
Gm5420
|
predicted gene 5420 |
| chr1_-_14755966 | 0.67 |
ENSMUST00000027062.5
|
Msc
|
musculin |
| chr8_-_69089200 | 0.67 |
ENSMUST00000037478.6
|
Slc18a1
|
solute carrier family 18 (vesicular monoamine), member 1 |
| chr15_-_99457742 | 0.67 |
ENSMUST00000023747.7
|
Nckap5l
|
NCK-associated protein 5-like |
| chr5_-_124249758 | 0.67 |
ENSMUST00000162812.1
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr11_-_69900886 | 0.66 |
ENSMUST00000108621.2
ENSMUST00000100969.2 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr12_-_112929415 | 0.64 |
ENSMUST00000075827.3
|
Jag2
|
jagged 2 |
| chr3_-_95687846 | 0.64 |
ENSMUST00000015994.3
ENSMUST00000148854.1 ENSMUST00000117782.1 |
Adamtsl4
|
ADAMTS-like 4 |
| chr17_-_57087729 | 0.64 |
ENSMUST00000071135.5
|
Tubb4a
|
tubulin, beta 4A class IVA |
| chr1_+_135836380 | 0.64 |
ENSMUST00000178204.1
|
Tnnt2
|
troponin T2, cardiac |
| chr1_+_134182404 | 0.64 |
ENSMUST00000153856.1
ENSMUST00000082060.3 ENSMUST00000133701.1 ENSMUST00000132873.1 |
Chi3l1
|
chitinase 3-like 1 |
| chr5_-_52566264 | 0.64 |
ENSMUST00000039750.5
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr1_+_87755870 | 0.63 |
ENSMUST00000144047.1
ENSMUST00000027512.6 ENSMUST00000113186.1 ENSMUST00000113190.2 |
Atg16l1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr8_-_84044982 | 0.63 |
ENSMUST00000061923.4
|
Rln3
|
relaxin 3 |
| chr4_-_118437331 | 0.63 |
ENSMUST00000006565.6
|
Cdc20
|
cell division cycle 20 |
| chr4_+_131921771 | 0.63 |
ENSMUST00000094666.3
|
Tmem200b
|
transmembrane protein 200B |
| chr9_+_15239045 | 0.62 |
ENSMUST00000034413.6
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr15_+_81811414 | 0.62 |
ENSMUST00000023024.7
|
Tef
|
thyrotroph embryonic factor |
| chr10_+_78574492 | 0.62 |
ENSMUST00000105384.3
|
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr18_+_34840575 | 0.62 |
ENSMUST00000043484.7
|
Reep2
|
receptor accessory protein 2 |
| chr5_+_33983437 | 0.62 |
ENSMUST00000114384.1
ENSMUST00000094869.5 ENSMUST00000114383.1 |
Gm1673
|
predicted gene 1673 |
| chr15_-_99651580 | 0.62 |
ENSMUST00000171908.1
ENSMUST00000171702.1 ENSMUST00000109581.2 ENSMUST00000169810.1 ENSMUST00000023756.5 |
Racgap1
|
Rac GTPase-activating protein 1 |
| chr10_+_79716588 | 0.62 |
ENSMUST00000099513.1
ENSMUST00000020581.2 |
Hcn2
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 2 |
| chr4_-_117883428 | 0.61 |
ENSMUST00000030266.5
|
B4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr11_-_120661175 | 0.61 |
ENSMUST00000150458.1
|
Notum
|
notum pectinacetylesterase homolog (Drosophila) |
| chr8_+_94532990 | 0.60 |
ENSMUST00000048653.2
ENSMUST00000109537.1 |
Cpne2
|
copine II |
| chr15_+_85736107 | 0.60 |
ENSMUST00000057979.5
|
Ppara
|
peroxisome proliferator activated receptor alpha |
| chr4_+_5644084 | 0.60 |
ENSMUST00000054857.6
|
Fam110b
|
family with sequence similarity 110, member B |
| chr9_-_95845215 | 0.60 |
ENSMUST00000093800.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr8_-_117673682 | 0.60 |
ENSMUST00000173522.1
ENSMUST00000174450.1 |
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr8_+_84148025 | 0.59 |
ENSMUST00000143833.1
ENSMUST00000118856.1 |
4930432K21Rik
|
RIKEN cDNA 4930432K21 gene |
| chr18_+_53465051 | 0.59 |
ENSMUST00000154557.1
|
Prdm6
|
PR domain containing 6 |
| chr6_-_87981482 | 0.59 |
ENSMUST00000056403.5
|
H1fx
|
H1 histone family, member X |
| chr11_-_69900930 | 0.59 |
ENSMUST00000018714.6
ENSMUST00000128046.1 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr7_+_5051515 | 0.59 |
ENSMUST00000069324.5
|
Zfp580
|
zinc finger protein 580 |
| chr2_+_70474923 | 0.59 |
ENSMUST00000100043.2
|
Sp5
|
trans-acting transcription factor 5 |
| chr14_-_18894255 | 0.58 |
ENSMUST00000124353.1
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr11_-_77489666 | 0.58 |
ENSMUST00000037593.7
ENSMUST00000092892.3 |
Ankrd13b
|
ankyrin repeat domain 13b |
| chr1_-_133906973 | 0.58 |
ENSMUST00000126123.1
|
Optc
|
opticin |
| chr2_-_10130638 | 0.58 |
ENSMUST00000042290.7
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr9_+_57504012 | 0.57 |
ENSMUST00000080514.7
|
Rpp25
|
ribonuclease P/MRP 25 subunit |
| chr11_+_71750980 | 0.57 |
ENSMUST00000108511.1
|
Wscd1
|
WSC domain containing 1 |
| chr9_+_44043384 | 0.57 |
ENSMUST00000114840.1
|
Thy1
|
thymus cell antigen 1, theta |
| chr6_-_18030435 | 0.57 |
ENSMUST00000010941.2
|
Wnt2
|
wingless-related MMTV integration site 2 |
| chr8_+_84148252 | 0.57 |
ENSMUST00000093375.4
|
4930432K21Rik
|
RIKEN cDNA 4930432K21 gene |
| chr15_+_102966794 | 0.57 |
ENSMUST00000001699.7
|
Hoxc10
|
homeobox C10 |
| chrX_+_160390684 | 0.56 |
ENSMUST00000112408.2
ENSMUST00000112402.1 ENSMUST00000112401.1 ENSMUST00000112400.1 ENSMUST00000112405.2 ENSMUST00000112404.2 ENSMUST00000146805.1 |
Gpr64
|
G protein-coupled receptor 64 |
| chr1_-_119053339 | 0.56 |
ENSMUST00000161301.1
ENSMUST00000161451.1 ENSMUST00000162607.1 |
Gli2
|
GLI-Kruppel family member GLI2 |
| chr7_+_45335256 | 0.56 |
ENSMUST00000085351.4
|
Hrc
|
histidine rich calcium binding protein |
| chr19_-_5106967 | 0.55 |
ENSMUST00000025804.5
|
Rab1b
|
RAB1B, member RAS oncogene family |
| chr7_+_24897381 | 0.55 |
ENSMUST00000003469.7
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha) |
| chr11_+_70647258 | 0.55 |
ENSMUST00000037534.7
|
Rnf167
|
ring finger protein 167 |
| chr5_-_35679416 | 0.55 |
ENSMUST00000114233.2
|
Htra3
|
HtrA serine peptidase 3 |
| chr5_+_110544326 | 0.54 |
ENSMUST00000040001.7
|
Galnt9
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 |
| chr7_-_44670820 | 0.54 |
ENSMUST00000048102.7
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr1_-_75505641 | 0.54 |
ENSMUST00000155084.1
|
Obsl1
|
obscurin-like 1 |
| chr11_+_82388900 | 0.54 |
ENSMUST00000054245.4
ENSMUST00000092852.2 |
Tmem132e
|
transmembrane protein 132E |
| chr5_+_145345254 | 0.54 |
ENSMUST00000079268.7
|
Cyp3a57
|
cytochrome P450, family 3, subfamily a, polypeptide 57 |
| chr10_-_81266906 | 0.53 |
ENSMUST00000046114.4
|
Mrpl54
|
mitochondrial ribosomal protein L54 |
| chr4_+_62965560 | 0.53 |
ENSMUST00000030043.6
ENSMUST00000107415.1 ENSMUST00000064814.5 |
Zfp618
|
zinc finger protein 618 |
| chr19_-_39812744 | 0.53 |
ENSMUST00000162507.1
ENSMUST00000160476.1 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr5_-_131538687 | 0.53 |
ENSMUST00000161374.1
|
Auts2
|
autism susceptibility candidate 2 |
| chr7_+_126272589 | 0.53 |
ENSMUST00000056028.9
|
Sbk1
|
SH3-binding kinase 1 |
| chr16_+_33062512 | 0.52 |
ENSMUST00000023497.2
|
Lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr10_-_26373956 | 0.52 |
ENSMUST00000105519.3
ENSMUST00000040219.6 |
L3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chrX_+_150594420 | 0.52 |
ENSMUST00000112713.2
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr17_+_12584183 | 0.52 |
ENSMUST00000046959.7
|
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr12_-_100899436 | 0.52 |
ENSMUST00000053668.3
|
Gpr68
|
G protein-coupled receptor 68 |
| chr2_+_158306493 | 0.52 |
ENSMUST00000016168.2
ENSMUST00000109491.1 |
Lbp
|
lipopolysaccharide binding protein |
| chr2_-_113829069 | 0.51 |
ENSMUST00000024005.7
|
Scg5
|
secretogranin V |
| chr2_+_155381808 | 0.51 |
ENSMUST00000043237.7
ENSMUST00000174685.1 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr7_+_43408187 | 0.51 |
ENSMUST00000005592.6
|
Siglecg
|
sialic acid binding Ig-like lectin G |
| chr19_-_42431778 | 0.51 |
ENSMUST00000048630.6
|
Crtac1
|
cartilage acidic protein 1 |
| chr11_-_12026732 | 0.51 |
ENSMUST00000143915.1
|
Grb10
|
growth factor receptor bound protein 10 |
| chr2_-_26910569 | 0.51 |
ENSMUST00000015920.5
ENSMUST00000139815.1 ENSMUST00000102899.3 |
Med22
|
mediator complex subunit 22 |
| chr12_+_112489448 | 0.51 |
ENSMUST00000057465.6
|
A530016L24Rik
|
RIKEN cDNA A530016L24 gene |
| chr2_-_91636394 | 0.51 |
ENSMUST00000111335.1
ENSMUST00000028681.8 |
F2
|
coagulation factor II |
| chr7_-_4752972 | 0.51 |
ENSMUST00000183971.1
ENSMUST00000182173.1 ENSMUST00000182738.1 ENSMUST00000184143.1 ENSMUST00000182111.1 ENSMUST00000182048.1 ENSMUST00000063324.7 |
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr7_-_67803489 | 0.50 |
ENSMUST00000181235.1
|
4833412C05Rik
|
RIKEN cDNA 4833412C05 gene |
| chr2_-_102451792 | 0.50 |
ENSMUST00000099678.3
|
Fjx1
|
four jointed box 1 (Drosophila) |
| chr8_+_57455898 | 0.50 |
ENSMUST00000034023.3
|
Scrg1
|
scrapie responsive gene 1 |
| chr5_-_120795530 | 0.50 |
ENSMUST00000100785.3
|
Oas1e
|
2'-5' oligoadenylate synthetase 1E |
| chrX_-_7574120 | 0.49 |
ENSMUST00000045924.7
ENSMUST00000115742.2 ENSMUST00000150787.1 |
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F |
| chr17_-_46680870 | 0.49 |
ENSMUST00000165007.1
ENSMUST00000071841.5 |
Klhdc3
|
kelch domain containing 3 |
| chr11_+_69965396 | 0.49 |
ENSMUST00000018713.6
|
Cldn7
|
claudin 7 |
| chr9_-_108263887 | 0.49 |
ENSMUST00000166905.1
ENSMUST00000080435.2 |
Dag1
|
dystroglycan 1 |
| chr1_-_127677923 | 0.49 |
ENSMUST00000160616.1
|
Tmem163
|
transmembrane protein 163 |
| chr11_+_121702591 | 0.49 |
ENSMUST00000125580.1
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr4_-_149137536 | 0.49 |
ENSMUST00000176124.1
ENSMUST00000177408.1 ENSMUST00000105695.1 ENSMUST00000030813.3 |
Apitd1
|
apoptosis-inducing, TAF9-like domain 1 |
| chr2_+_27886416 | 0.49 |
ENSMUST00000028280.7
|
Col5a1
|
collagen, type V, alpha 1 |
| chr16_-_30310773 | 0.49 |
ENSMUST00000061190.6
|
Gp5
|
glycoprotein 5 (platelet) |
| chr6_-_118197732 | 0.49 |
ENSMUST00000032201.5
ENSMUST00000088790.3 |
Ret
|
ret proto-oncogene |
| chr11_-_114795888 | 0.48 |
ENSMUST00000000206.3
|
Btbd17
|
BTB (POZ) domain containing 17 |
| chr9_+_103112072 | 0.48 |
ENSMUST00000035155.6
|
Rab6b
|
RAB6B, member RAS oncogene family |
| chr14_+_31217850 | 0.48 |
ENSMUST00000090180.2
|
Sema3g
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| chr2_-_165388245 | 0.48 |
ENSMUST00000103084.3
|
Zfp334
|
zinc finger protein 334 |
| chr5_+_90772435 | 0.48 |
ENSMUST00000031320.6
|
Pf4
|
platelet factor 4 |
| chr17_+_46681038 | 0.48 |
ENSMUST00000002845.6
|
Mea1
|
male enhanced antigen 1 |
| chr7_+_79053483 | 0.47 |
ENSMUST00000032835.5
|
Acan
|
aggrecan |
| chr9_-_62510498 | 0.47 |
ENSMUST00000164246.2
|
Coro2b
|
coronin, actin binding protein, 2B |
| chr1_-_75506331 | 0.47 |
ENSMUST00000113567.2
ENSMUST00000113565.2 |
Obsl1
|
obscurin-like 1 |
| chr8_+_13435459 | 0.47 |
ENSMUST00000167071.1
ENSMUST00000167505.1 |
Tmem255b
|
transmembrane protein 255B |
| chr2_-_92392634 | 0.46 |
ENSMUST00000111279.2
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr16_+_17646564 | 0.46 |
ENSMUST00000182117.1
ENSMUST00000182671.1 ENSMUST00000182344.1 |
Ccdc74a
|
coiled-coil domain containing 74A |
| chr6_-_83656082 | 0.45 |
ENSMUST00000014686.2
|
Clec4f
|
C-type lectin domain family 4, member f |
| chr9_-_121916288 | 0.45 |
ENSMUST00000062474.4
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr17_-_46890405 | 0.45 |
ENSMUST00000086675.3
|
A330017A19Rik
|
RIKEN cDNA A330017A19 gene |
| chr11_-_102365111 | 0.45 |
ENSMUST00000006749.9
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gcm2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.9 | 0.9 | GO:0048389 | intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.5 | 1.6 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.5 | 3.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 | 0.3 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.3 | 1.6 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.3 | 1.2 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.3 | 4.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 1.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.3 | 0.8 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.3 | 1.3 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.3 | 0.8 | GO:0060752 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.3 | 1.0 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.2 | 0.7 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.2 | 1.4 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 | 0.9 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 0.6 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 0.6 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 0.2 | 0.6 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.2 | 0.6 | GO:0006507 | GPI anchor release(GO:0006507) |
| 0.2 | 0.8 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.2 | 1.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 1.5 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.2 | 0.6 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.2 | 2.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 0.7 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.2 | 0.5 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 | 0.5 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.2 | 0.7 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 | 0.7 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.6 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 0.2 | 2.0 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.2 | 0.3 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.6 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.4 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 1.9 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.3 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 0.1 | 0.3 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 0.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 1.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.6 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 1.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 0.7 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.4 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.4 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.4 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.7 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 2.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.5 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.5 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.6 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.1 | 0.2 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.1 | 0.4 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.7 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.6 | GO:0072363 | regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 0.1 | 2.7 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 1.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.4 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.1 | 0.4 | GO:0072166 | posterior mesonephric tubule development(GO:0072166) metanephric comma-shaped body morphogenesis(GO:0072278) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.1 | 0.3 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.8 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.3 | GO:1905204 | negative regulation of connective tissue replacement(GO:1905204) positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.1 | 0.8 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.3 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) |
| 0.1 | 0.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.6 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.3 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.1 | 0.4 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.4 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.3 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.7 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.5 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.7 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.6 | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.1 | 0.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.3 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.1 | 0.2 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 0.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.4 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.1 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 0.3 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 | 0.1 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.1 | 0.3 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.2 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.1 | 0.3 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.3 | GO:0021666 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.1 | 0.3 | GO:0098912 | membrane depolarization during AV node cell action potential(GO:0086045) membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.1 | 0.5 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.2 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.1 | 0.4 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.1 | 0.3 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 0.9 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.4 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.2 | GO:0050883 | regulation of sulfur amino acid metabolic process(GO:0031335) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 | 0.7 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.8 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.1 | 10.3 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 1.1 | GO:0086027 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.1 | 0.2 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.2 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.1 | 0.2 | GO:0015881 | creatine transport(GO:0015881) |
| 0.1 | 0.3 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 2.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.9 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.4 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.1 | 0.3 | GO:0002587 | negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.1 | 0.5 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 | 0.4 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.5 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.2 | GO:0014064 | positive regulation of serotonin secretion(GO:0014064) sperm ejaculation(GO:0042713) |
| 0.1 | 0.2 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 | 0.3 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.6 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 1.6 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.1 | 0.9 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.1 | 0.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.4 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.2 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.4 | GO:0032796 | uropod organization(GO:0032796) |
| 0.1 | 0.4 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.4 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 0.2 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.9 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 1.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 0.2 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.1 | 0.5 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.1 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) |
| 0.1 | 0.6 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 0.2 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 0.1 | 0.3 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 0.2 | GO:0006532 | aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.6 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.9 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.5 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.1 | 0.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 1.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.3 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0043091 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.7 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 1.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.0 | 0.2 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.8 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 1.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.2 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.3 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.7 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.2 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.0 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 1.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.3 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil activation(GO:0043307) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.0 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 0.2 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.2 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.4 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.6 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.3 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.6 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.5 | GO:0051608 | histamine transport(GO:0051608) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.4 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.4 | GO:0031111 | negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.3 | GO:0015670 | gas transport(GO:0015669) carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.4 | GO:1902572 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.0 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.8 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 1.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.2 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.2 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.7 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.2 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.3 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.2 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.7 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.7 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.7 | GO:0045823 | positive regulation of heart contraction(GO:0045823) |
| 0.0 | 0.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.4 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.6 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.7 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 1.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0046968 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) peptide antigen transport(GO:0046968) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:2000795 | lung ciliated cell differentiation(GO:0061141) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.2 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 1.5 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 1.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.2 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.2 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.1 | GO:1903632 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.0 | GO:0046122 | deoxyribonucleoside catabolic process(GO:0046121) purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.9 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.3 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.2 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.2 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.4 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.1 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.2 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.0 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 1.3 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.2 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.0 | 0.4 | GO:0032272 | negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.0 | 0.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.2 | GO:1903020 | positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.2 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.2 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.0 | 0.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.1 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.3 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.0 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.0 | 0.0 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.0 | GO:1905035 | regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.0 | 0.1 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.6 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.2 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.0 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.3 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.2 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.0 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.0 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.3 | 1.3 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.3 | 0.8 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.2 | 1.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.2 | 0.6 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 1.9 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 0.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 0.7 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.8 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 0.5 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.2 | 0.5 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.1 | 0.7 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.6 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.4 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 1.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 1.1 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.5 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.6 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 2.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 1.2 | GO:0042571 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.2 | GO:0005940 | septin ring(GO:0005940) |
| 0.1 | 0.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.7 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 0.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.2 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.1 | 3.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 0.3 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.5 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.9 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 6.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 1.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 6.3 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 19.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.9 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.5 | 1.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.4 | 1.7 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.3 | 0.8 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 1.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.2 | 0.7 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.2 | 0.7 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 1.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 1.6 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.2 | 0.9 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 0.6 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 0.5 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.2 | 0.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 2.1 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 1.0 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 1.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 1.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.4 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.1 | 0.4 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 5.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.4 | GO:0045352 | interleukin-1, Type II receptor binding(GO:0005151) interleukin-1 Type I receptor antagonist activity(GO:0045352) interleukin-1 Type II receptor antagonist activity(GO:0045353) |
| 0.1 | 0.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.4 | GO:0004923 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.1 | 1.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.7 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.7 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.5 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.1 | 0.4 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.3 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.1 | 0.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.7 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.8 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 0.3 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.3 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.6 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 18.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.3 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.1 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.5 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.3 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.7 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 0.4 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.4 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.2 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.1 | 1.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 0.2 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.1 | 2.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.3 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 0.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.2 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.1 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 2.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.5 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 1.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 2.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.5 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.5 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.3 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.4 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.5 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.3 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 0.2 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 0.2 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.1 | 1.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.8 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.3 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 2.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.2 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 1.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 2.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.9 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.4 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.5 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 3.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.7 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 3.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0060229 | lipase activator activity(GO:0060229) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.3 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.2 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.0 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 0.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 11.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 5.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.0 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 7.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 0.2 | REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | Genes involved in Negative regulation of FGFR signaling |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.1 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.1 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 2.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 2.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME SIGNALLING TO RAS | Genes involved in Signalling to RAS |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |