Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Foxk1_Foxj1
Z-value: 0.85
Transcription factors associated with Foxk1_Foxj1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
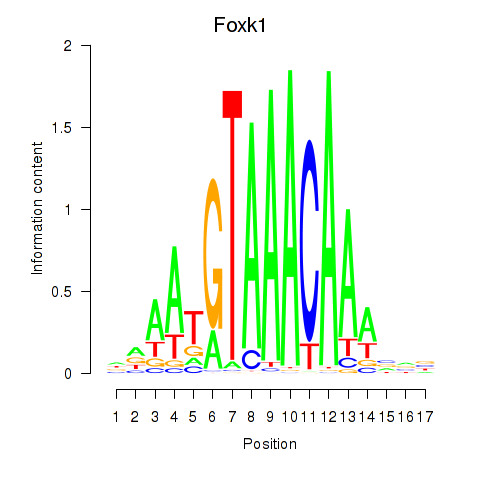
Foxk1
|
ENSMUSG00000056493.8 | forkhead box K1 |
|
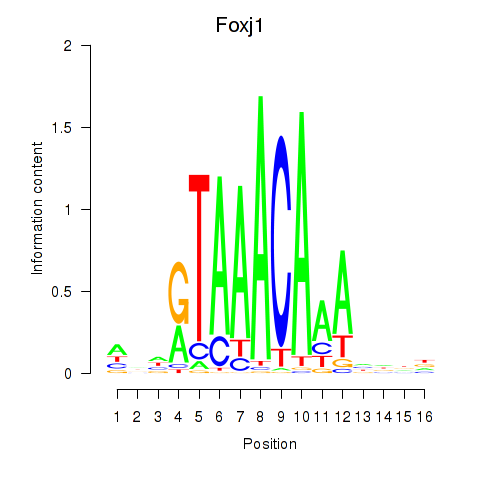
Foxj1
|
ENSMUSG00000034227.7 | forkhead box J1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxj1 | mm10_v2_chr11_-_116335384_116335399 | 0.61 | 8.6e-05 | Click! |
| Foxk1 | mm10_v2_chr5_+_142401484_142401532 | 0.55 | 4.7e-04 | Click! |
Activity profile of Foxk1_Foxj1 motif
Sorted Z-values of Foxk1_Foxj1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_106167564 | 11.74 |
ENSMUST00000063062.8
|
Chi3l3
|
chitinase 3-like 3 |
| chr13_-_4150628 | 7.09 |
ENSMUST00000110704.2
ENSMUST00000021635.7 |
Akr1c18
|
aldo-keto reductase family 1, member C18 |
| chrX_-_162964557 | 5.69 |
ENSMUST00000038769.2
|
S100g
|
S100 calcium binding protein G |
| chr14_-_70630149 | 4.67 |
ENSMUST00000022694.9
|
Dmtn
|
dematin actin binding protein |
| chr6_+_34412334 | 4.29 |
ENSMUST00000007449.8
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr17_+_40811089 | 4.18 |
ENSMUST00000024721.7
|
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr1_+_131638485 | 4.01 |
ENSMUST00000112411.1
|
Ctse
|
cathepsin E |
| chr4_-_117178726 | 3.72 |
ENSMUST00000153953.1
ENSMUST00000106436.1 |
Kif2c
|
kinesin family member 2C |
| chr3_-_113532288 | 3.56 |
ENSMUST00000132353.1
|
Amy2a1
|
amylase 2a1 |
| chr2_+_174450678 | 3.44 |
ENSMUST00000016399.5
|
Tubb1
|
tubulin, beta 1 class VI |
| chr6_-_83536215 | 3.21 |
ENSMUST00000075161.5
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr4_-_44710408 | 3.15 |
ENSMUST00000134968.2
ENSMUST00000173821.1 ENSMUST00000174319.1 ENSMUST00000173733.1 ENSMUST00000172866.1 ENSMUST00000165417.2 ENSMUST00000107825.2 ENSMUST00000102932.3 ENSMUST00000107827.2 ENSMUST00000107826.2 |
Pax5
|
paired box gene 5 |
| chr19_+_58759700 | 3.09 |
ENSMUST00000026081.3
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chrX_-_136203637 | 3.08 |
ENSMUST00000151592.1
ENSMUST00000131510.1 ENSMUST00000066819.4 |
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr3_+_159495408 | 2.63 |
ENSMUST00000120272.1
ENSMUST00000029825.7 ENSMUST00000106041.2 |
Depdc1a
|
DEP domain containing 1a |
| chr5_-_145805862 | 2.55 |
ENSMUST00000067479.5
|
Cyp3a44
|
cytochrome P450, family 3, subfamily a, polypeptide 44 |
| chr4_+_44943727 | 2.54 |
ENSMUST00000154177.1
|
Gm12678
|
predicted gene 12678 |
| chr6_-_40999479 | 2.52 |
ENSMUST00000166306.1
|
Gm2663
|
predicted gene 2663 |
| chr7_+_26061495 | 2.43 |
ENSMUST00000005669.7
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr17_-_23645264 | 2.35 |
ENSMUST00000024696.7
|
Mmp25
|
matrix metallopeptidase 25 |
| chr4_-_87806296 | 2.24 |
ENSMUST00000126353.1
ENSMUST00000149357.1 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr3_+_51661167 | 2.21 |
ENSMUST00000099106.3
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr7_+_26173411 | 2.17 |
ENSMUST00000082214.4
|
Cyp2b9
|
cytochrome P450, family 2, subfamily b, polypeptide 9 |
| chr1_+_57774600 | 2.11 |
ENSMUST00000167971.1
ENSMUST00000170139.1 ENSMUST00000171699.1 ENSMUST00000164302.1 |
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr15_+_85510812 | 2.04 |
ENSMUST00000079690.2
|
Gm4825
|
predicted pseudogene 4825 |
| chr7_-_101869307 | 2.03 |
ENSMUST00000140584.1
ENSMUST00000134145.1 |
Folr1
|
folate receptor 1 (adult) |
| chr3_+_51661209 | 2.00 |
ENSMUST00000161590.1
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr13_-_51734695 | 1.98 |
ENSMUST00000110039.1
|
Sema4d
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr12_+_111971545 | 1.96 |
ENSMUST00000079009.5
|
Tdrd9
|
tudor domain containing 9 |
| chr1_+_136467958 | 1.92 |
ENSMUST00000047817.6
|
Kif14
|
kinesin family member 14 |
| chr3_-_20275659 | 1.87 |
ENSMUST00000011607.5
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr1_+_57774842 | 1.84 |
ENSMUST00000167085.1
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr6_+_8520008 | 1.81 |
ENSMUST00000162567.1
ENSMUST00000161217.1 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chr17_+_29114142 | 1.81 |
ENSMUST00000141797.1
ENSMUST00000132262.1 ENSMUST00000141239.1 ENSMUST00000138816.1 |
Gm16194
|
predicted gene 16194 |
| chr2_-_170194033 | 1.78 |
ENSMUST00000180625.1
|
Gm17619
|
predicted gene, 17619 |
| chr19_-_42086338 | 1.71 |
ENSMUST00000051772.8
|
Morn4
|
MORN repeat containing 4 |
| chr1_+_107535508 | 1.69 |
ENSMUST00000182198.1
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr4_-_87806276 | 1.68 |
ENSMUST00000148059.1
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr3_+_153844209 | 1.67 |
ENSMUST00000044089.3
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr15_-_103255433 | 1.67 |
ENSMUST00000075192.6
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr4_+_150927918 | 1.66 |
ENSMUST00000139826.1
ENSMUST00000116257.1 |
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chrX_-_104671048 | 1.60 |
ENSMUST00000042070.5
|
Zdhhc15
|
zinc finger, DHHC domain containing 15 |
| chr7_+_45554893 | 1.54 |
ENSMUST00000107752.3
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr8_-_53638945 | 1.52 |
ENSMUST00000047768.4
|
Neil3
|
nei like 3 (E. coli) |
| chr16_-_76022266 | 1.51 |
ENSMUST00000114240.1
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr16_-_22161450 | 1.51 |
ENSMUST00000115379.1
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr19_+_37376359 | 1.49 |
ENSMUST00000012587.3
|
Kif11
|
kinesin family member 11 |
| chr19_-_34255325 | 1.49 |
ENSMUST00000039631.8
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr2_-_118728345 | 1.46 |
ENSMUST00000159756.1
|
Plcb2
|
phospholipase C, beta 2 |
| chr14_-_57826128 | 1.44 |
ENSMUST00000022536.2
|
Ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr14_+_11227511 | 1.44 |
ENSMUST00000080237.3
|
Rpl21-ps4
|
ribosomal protein L21, pseudogene 4 |
| chr1_-_144775419 | 1.44 |
ENSMUST00000027603.3
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr6_-_136941887 | 1.43 |
ENSMUST00000111891.1
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr7_-_142699510 | 1.43 |
ENSMUST00000105934.1
|
Ins2
|
insulin II |
| chr9_+_110344185 | 1.43 |
ENSMUST00000142100.1
|
Scap
|
SREBF chaperone |
| chr16_+_48872608 | 1.42 |
ENSMUST00000065666.4
|
Retnlg
|
resistin like gamma |
| chr6_+_78425973 | 1.42 |
ENSMUST00000079926.5
|
Reg1
|
regenerating islet-derived 1 |
| chr11_+_58948890 | 1.40 |
ENSMUST00000078267.3
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr5_-_44099220 | 1.37 |
ENSMUST00000165909.1
|
Prom1
|
prominin 1 |
| chr17_+_5799491 | 1.35 |
ENSMUST00000181484.1
|
3300005D01Rik
|
RIKEN cDNA 3300005D01 gene |
| chr6_-_129917650 | 1.31 |
ENSMUST00000118060.1
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr10_-_97726755 | 1.30 |
ENSMUST00000166373.1
|
C030005K15Rik
|
RIKEN cDNA C030005K15 gene |
| chr19_-_8218832 | 1.29 |
ENSMUST00000113298.2
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr16_+_44943678 | 1.27 |
ENSMUST00000114613.2
ENSMUST00000114612.1 ENSMUST00000077178.6 ENSMUST00000048479.7 ENSMUST00000114611.3 ENSMUST00000164007.1 ENSMUST00000171779.1 |
Cd200r3
|
CD200 receptor 3 |
| chr13_-_22219820 | 1.27 |
ENSMUST00000057516.1
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr2_+_119047116 | 1.27 |
ENSMUST00000152380.1
ENSMUST00000099542.2 |
Casc5
|
cancer susceptibility candidate 5 |
| chr4_-_141825997 | 1.26 |
ENSMUST00000102481.3
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr14_-_61258484 | 1.25 |
ENSMUST00000077954.5
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr15_-_78855517 | 1.24 |
ENSMUST00000044584.4
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr4_+_11191354 | 1.18 |
ENSMUST00000170901.1
|
Ccne2
|
cyclin E2 |
| chr1_-_45503282 | 1.17 |
ENSMUST00000086430.4
|
Col5a2
|
collagen, type V, alpha 2 |
| chr3_-_27896360 | 1.17 |
ENSMUST00000058077.3
|
Tmem212
|
transmembrane protein 212 |
| chr3_-_65529355 | 1.16 |
ENSMUST00000099076.3
|
4931440P22Rik
|
RIKEN cDNA 4931440P22 gene |
| chr4_-_43499608 | 1.16 |
ENSMUST00000136005.1
ENSMUST00000054538.6 |
Arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr15_+_6579841 | 1.16 |
ENSMUST00000090461.5
|
Fyb
|
FYN binding protein |
| chr17_+_47726834 | 1.15 |
ENSMUST00000024782.5
ENSMUST00000144955.1 |
Pgc
|
progastricsin (pepsinogen C) |
| chr5_+_30666886 | 1.14 |
ENSMUST00000144742.1
|
Cenpa
|
centromere protein A |
| chr14_+_55765956 | 1.14 |
ENSMUST00000057569.3
|
Ltb4r1
|
leukotriene B4 receptor 1 |
| chr9_-_58741543 | 1.10 |
ENSMUST00000098674.4
|
2410076I21Rik
|
RIKEN cDNA 2410076I21 gene |
| chr5_+_150522599 | 1.08 |
ENSMUST00000044620.7
|
Brca2
|
breast cancer 2 |
| chr6_-_122609964 | 1.08 |
ENSMUST00000032211.4
|
Gdf3
|
growth differentiation factor 3 |
| chr4_-_154928545 | 1.07 |
ENSMUST00000152687.1
|
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr15_-_36879816 | 1.07 |
ENSMUST00000100713.2
|
Gm10384
|
predicted gene 10384 |
| chr11_+_11487671 | 1.06 |
ENSMUST00000020410.4
|
4930415F15Rik
|
RIKEN cDNA 4930415F15 gene |
| chr5_+_136038496 | 1.03 |
ENSMUST00000062606.6
|
Upk3b
|
uroplakin 3B |
| chr18_-_23981555 | 1.01 |
ENSMUST00000115829.1
|
Zscan30
|
zinc finger and SCAN domain containing 30 |
| chr3_+_84666192 | 1.01 |
ENSMUST00000107682.1
|
Tmem154
|
transmembrane protein 154 |
| chr7_+_24897381 | 1.01 |
ENSMUST00000003469.7
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha) |
| chr10_-_62792243 | 1.00 |
ENSMUST00000020268.5
|
Ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr2_+_119047129 | 0.99 |
ENSMUST00000153300.1
ENSMUST00000028799.5 |
Casc5
|
cancer susceptibility candidate 5 |
| chr17_-_25944932 | 0.97 |
ENSMUST00000085027.3
|
Nhlrc4
|
NHL repeat containing 4 |
| chr3_+_67374116 | 0.96 |
ENSMUST00000061322.8
|
Mlf1
|
myeloid leukemia factor 1 |
| chr6_+_42264983 | 0.96 |
ENSMUST00000031895.6
|
Casp2
|
caspase 2 |
| chr10_+_115569986 | 0.95 |
ENSMUST00000173620.1
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr11_+_46235460 | 0.94 |
ENSMUST00000060185.2
|
Fndc9
|
fibronectin type III domain containing 9 |
| chr19_-_38043559 | 0.94 |
ENSMUST00000041475.8
ENSMUST00000172095.2 |
Myof
|
myoferlin |
| chr18_+_21072329 | 0.93 |
ENSMUST00000082235.4
|
Mep1b
|
meprin 1 beta |
| chr4_-_116167591 | 0.93 |
ENSMUST00000030465.3
ENSMUST00000143426.1 |
Tspan1
|
tetraspanin 1 |
| chr16_-_22657182 | 0.93 |
ENSMUST00000023578.7
|
Dgkg
|
diacylglycerol kinase, gamma |
| chrX_+_170010744 | 0.92 |
ENSMUST00000178789.1
|
Gm21887
|
predicted gene, 21887 |
| chr8_+_93810832 | 0.91 |
ENSMUST00000034198.8
ENSMUST00000125716.1 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr4_+_154869585 | 0.90 |
ENSMUST00000079269.7
ENSMUST00000163732.1 ENSMUST00000080559.6 |
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr13_-_90905321 | 0.90 |
ENSMUST00000109541.3
|
Atp6ap1l
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr3_-_59210881 | 0.90 |
ENSMUST00000040622.1
|
P2ry13
|
purinergic receptor P2Y, G-protein coupled 13 |
| chr17_-_28689987 | 0.90 |
ENSMUST00000114764.1
|
Slc26a8
|
solute carrier family 26, member 8 |
| chr9_+_70678950 | 0.89 |
ENSMUST00000067880.6
|
Adam10
|
a disintegrin and metallopeptidase domain 10 |
| chr16_+_38362205 | 0.89 |
ENSMUST00000023494.6
|
Popdc2
|
popeye domain containing 2 |
| chr3_-_101287897 | 0.89 |
ENSMUST00000029456.4
|
Cd2
|
CD2 antigen |
| chr3_-_100969644 | 0.87 |
ENSMUST00000076941.5
|
Ttf2
|
transcription termination factor, RNA polymerase II |
| chr2_-_84822546 | 0.87 |
ENSMUST00000028471.5
|
Smtnl1
|
smoothelin-like 1 |
| chr19_-_20390944 | 0.87 |
ENSMUST00000025561.7
|
Anxa1
|
annexin A1 |
| chr1_+_153749414 | 0.86 |
ENSMUST00000086209.3
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr1_+_164062070 | 0.86 |
ENSMUST00000097491.3
ENSMUST00000027871.7 |
Sell
|
selectin, lymphocyte |
| chr3_-_101287879 | 0.86 |
ENSMUST00000152321.1
|
Cd2
|
CD2 antigen |
| chr1_+_179546303 | 0.85 |
ENSMUST00000040706.8
|
Cnst
|
consortin, connexin sorting protein |
| chr11_+_101733011 | 0.85 |
ENSMUST00000129741.1
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr4_-_62470868 | 0.85 |
ENSMUST00000135811.1
ENSMUST00000120095.1 ENSMUST00000030087.7 ENSMUST00000107452.1 ENSMUST00000155522.1 |
Wdr31
|
WD repeat domain 31 |
| chr10_-_51631458 | 0.84 |
ENSMUST00000020062.3
|
Gprc6a
|
G protein-coupled receptor, family C, group 6, member A |
| chr3_+_67374091 | 0.84 |
ENSMUST00000077916.5
|
Mlf1
|
myeloid leukemia factor 1 |
| chr17_-_31144271 | 0.84 |
ENSMUST00000024826.7
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr10_+_127677064 | 0.84 |
ENSMUST00000118612.1
ENSMUST00000048099.4 |
Tmem194
|
transmembrane protein 194 |
| chr11_+_87755567 | 0.83 |
ENSMUST00000123700.1
|
A430104N18Rik
|
RIKEN cDNA A430104N18 gene |
| chr2_-_152831665 | 0.83 |
ENSMUST00000156688.1
ENSMUST00000007803.5 |
Bcl2l1
|
BCL2-like 1 |
| chrY_+_90785442 | 0.83 |
ENSMUST00000177591.1
ENSMUST00000177671.1 ENSMUST00000179077.1 |
Erdr1
|
erythroid differentiation regulator 1 |
| chr17_-_7827289 | 0.83 |
ENSMUST00000167580.1
ENSMUST00000169126.1 |
Fndc1
|
fibronectin type III domain containing 1 |
| chrX_+_9885622 | 0.81 |
ENSMUST00000067529.2
ENSMUST00000086165.3 |
Sytl5
|
synaptotagmin-like 5 |
| chr17_-_55915870 | 0.81 |
ENSMUST00000074828.4
|
Rpl21-ps6
|
ribosomal protein L21, pseudogene 6 |
| chr1_+_40324570 | 0.80 |
ENSMUST00000095020.3
|
Il1rl2
|
interleukin 1 receptor-like 2 |
| chr11_+_46810792 | 0.80 |
ENSMUST00000068877.6
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr10_+_88091070 | 0.80 |
ENSMUST00000048621.7
|
Pmch
|
pro-melanin-concentrating hormone |
| chr16_+_17146937 | 0.80 |
ENSMUST00000115706.1
ENSMUST00000069064.4 |
Ydjc
|
YdjC homolog (bacterial) |
| chr6_-_129507107 | 0.79 |
ENSMUST00000183258.1
ENSMUST00000182784.1 ENSMUST00000032265.6 ENSMUST00000162815.1 |
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr12_-_15816762 | 0.79 |
ENSMUST00000020922.7
|
Trib2
|
tribbles homolog 2 (Drosophila) |
| chr7_-_26939377 | 0.78 |
ENSMUST00000170227.1
|
Cyp2a22
|
cytochrome P450, family 2, subfamily a, polypeptide 22 |
| chr11_-_34833631 | 0.77 |
ENSMUST00000093191.2
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr5_+_121220191 | 0.75 |
ENSMUST00000119892.2
ENSMUST00000042614.6 |
Gm15800
|
predicted gene 15800 |
| chr19_+_53460610 | 0.75 |
ENSMUST00000180442.1
|
4833407H14Rik
|
RIKEN cDNA 4833407H14 gene |
| chr16_-_45492962 | 0.75 |
ENSMUST00000114585.2
|
Gm609
|
predicted gene 609 |
| chr18_+_67800101 | 0.75 |
ENSMUST00000025425.5
|
Cep192
|
centrosomal protein 192 |
| chr16_+_38362234 | 0.75 |
ENSMUST00000114739.1
|
Popdc2
|
popeye domain containing 2 |
| chr10_-_13324160 | 0.73 |
ENSMUST00000105545.4
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr16_-_20425881 | 0.72 |
ENSMUST00000077867.3
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr6_-_129740484 | 0.71 |
ENSMUST00000050385.5
|
Klri2
|
killer cell lectin-like receptor family I member 2 |
| chr17_+_28530834 | 0.71 |
ENSMUST00000025060.2
|
Armc12
|
armadillo repeat containing 12 |
| chr16_-_22657165 | 0.71 |
ENSMUST00000089925.3
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr8_+_75109528 | 0.71 |
ENSMUST00000164309.1
|
Mcm5
|
minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae) |
| chr17_+_9422060 | 0.71 |
ENSMUST00000076982.6
|
Gm17728
|
predicted gene, 17728 |
| chr16_-_15637277 | 0.70 |
ENSMUST00000023353.3
|
Mcm4
|
minichromosome maintenance deficient 4 homolog (S. cerevisiae) |
| chr12_-_40248073 | 0.70 |
ENSMUST00000169926.1
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chrX_+_163911401 | 0.70 |
ENSMUST00000140845.1
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr7_+_80186835 | 0.70 |
ENSMUST00000107383.1
ENSMUST00000032754.7 |
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr14_-_61258383 | 0.70 |
ENSMUST00000121148.1
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr13_+_76579670 | 0.70 |
ENSMUST00000126960.1
ENSMUST00000109583.2 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr7_+_19282613 | 0.69 |
ENSMUST00000032559.9
|
Rtn2
|
reticulon 2 (Z-band associated protein) |
| chr7_+_19291070 | 0.69 |
ENSMUST00000108468.3
|
Rtn2
|
reticulon 2 (Z-band associated protein) |
| chr7_-_6331235 | 0.69 |
ENSMUST00000127658.1
ENSMUST00000062765.7 |
Zfp583
|
zinc finger protein 583 |
| chrX_+_139217166 | 0.68 |
ENSMUST00000166444.1
ENSMUST00000170671.1 ENSMUST00000113041.2 ENSMUST00000113042.2 |
Mum1l1
|
melanoma associated antigen (mutated) 1-like 1 |
| chr2_-_73452666 | 0.67 |
ENSMUST00000151939.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chrX_+_56786527 | 0.67 |
ENSMUST00000144600.1
|
Fhl1
|
four and a half LIM domains 1 |
| chr5_+_122209729 | 0.67 |
ENSMUST00000072602.7
ENSMUST00000143560.1 |
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr16_-_36784784 | 0.66 |
ENSMUST00000165531.1
|
Slc15a2
|
solute carrier family 15 (H+/peptide transporter), member 2 |
| chr10_+_37139558 | 0.66 |
ENSMUST00000062667.3
|
5930403N24Rik
|
RIKEN cDNA 5930403N24 gene |
| chr18_-_43373248 | 0.66 |
ENSMUST00000118043.1
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr2_-_118728430 | 0.66 |
ENSMUST00000102524.1
|
Plcb2
|
phospholipase C, beta 2 |
| chr5_-_114823460 | 0.65 |
ENSMUST00000140374.1
ENSMUST00000100850.4 |
Gm20499
2610524H06Rik
|
predicted gene 20499 RIKEN cDNA 2610524H06 gene |
| chr16_-_4679703 | 0.65 |
ENSMUST00000038552.6
ENSMUST00000090480.4 |
Coro7
|
coronin 7 |
| chrX_+_107088452 | 0.65 |
ENSMUST00000150494.1
|
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr5_-_138172383 | 0.65 |
ENSMUST00000000505.9
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr11_+_11489266 | 0.64 |
ENSMUST00000109678.1
|
4930415F15Rik
|
RIKEN cDNA 4930415F15 gene |
| chr15_-_77022632 | 0.64 |
ENSMUST00000019037.8
ENSMUST00000169226.1 |
Mb
|
myoglobin |
| chr11_+_83065092 | 0.64 |
ENSMUST00000038038.7
|
Slfn2
|
schlafen 2 |
| chr4_+_102741287 | 0.64 |
ENSMUST00000097948.2
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr17_-_26099257 | 0.64 |
ENSMUST00000053575.3
|
Gm8186
|
predicted gene 8186 |
| chr13_-_19824234 | 0.64 |
ENSMUST00000065335.2
|
Gpr141
|
G protein-coupled receptor 141 |
| chr6_-_115037824 | 0.63 |
ENSMUST00000174848.1
ENSMUST00000032461.5 |
Tamm41
|
TAM41, mitochondrial translocator assembly and maintenance protein, homolog (S. cerevisiae) |
| chr7_+_26266831 | 0.63 |
ENSMUST00000057123.6
|
Vmn1r184
|
vomeronasal 1 receptor, 184 |
| chr7_+_126950837 | 0.62 |
ENSMUST00000106332.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr9_-_60511003 | 0.61 |
ENSMUST00000098660.3
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
| chr2_-_163645125 | 0.61 |
ENSMUST00000017851.3
|
Serinc3
|
serine incorporator 3 |
| chr15_-_75048837 | 0.61 |
ENSMUST00000179762.1
ENSMUST00000065408.9 |
Ly6c1
|
lymphocyte antigen 6 complex, locus C1 |
| chr6_-_69400097 | 0.61 |
ENSMUST00000177795.1
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chrX_+_166344692 | 0.61 |
ENSMUST00000112223.1
ENSMUST00000112224.1 ENSMUST00000112229.2 ENSMUST00000112228.1 ENSMUST00000112227.2 ENSMUST00000112226.2 |
Gpm6b
|
glycoprotein m6b |
| chr2_+_125152505 | 0.61 |
ENSMUST00000110494.2
ENSMUST00000028630.2 ENSMUST00000110495.2 |
Slc12a1
|
solute carrier family 12, member 1 |
| chr1_-_138175238 | 0.61 |
ENSMUST00000182536.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr10_-_30655859 | 0.60 |
ENSMUST00000092610.4
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr10_+_39420009 | 0.60 |
ENSMUST00000157009.1
|
Fyn
|
Fyn proto-oncogene |
| chr7_-_24299310 | 0.60 |
ENSMUST00000145131.1
|
Zfp61
|
zinc finger protein 61 |
| chr6_-_148896150 | 0.59 |
ENSMUST00000072324.5
ENSMUST00000111569.2 |
Caprin2
|
caprin family member 2 |
| chr5_+_110330697 | 0.59 |
ENSMUST00000112481.1
|
Pole
|
polymerase (DNA directed), epsilon |
| chr1_-_138175283 | 0.58 |
ENSMUST00000182755.1
ENSMUST00000183262.1 ENSMUST00000027645.7 ENSMUST00000112036.2 ENSMUST00000182283.1 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr5_+_139423151 | 0.58 |
ENSMUST00000066211.4
|
Gper1
|
G protein-coupled estrogen receptor 1 |
| chr11_+_101732950 | 0.58 |
ENSMUST00000039152.7
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr4_-_111902754 | 0.58 |
ENSMUST00000102719.1
ENSMUST00000102721.1 |
Slc5a9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| chr5_-_5749317 | 0.57 |
ENSMUST00000015796.2
|
Steap1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr10_+_17796256 | 0.57 |
ENSMUST00000037964.6
|
Txlnb
|
taxilin beta |
| chr19_-_24901309 | 0.57 |
ENSMUST00000058600.2
|
Foxd4
|
forkhead box D4 |
| chr10_+_97482350 | 0.56 |
ENSMUST00000163448.2
|
Dcn
|
decorin |
| chr5_+_114896936 | 0.56 |
ENSMUST00000031542.9
ENSMUST00000146072.1 ENSMUST00000150361.1 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr1_-_138175126 | 0.56 |
ENSMUST00000183301.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr14_+_26894557 | 0.56 |
ENSMUST00000090337.4
ENSMUST00000165929.2 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxk1_Foxj1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 1.6 | 4.7 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.4 | 4.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.7 | 3.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.6 | 1.9 | GO:0033624 | negative regulation of integrin activation(GO:0033624) |
| 0.5 | 2.0 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.5 | 1.4 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 0.5 | 1.4 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.4 | 1.7 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.4 | 0.4 | GO:2000388 | positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) |
| 0.4 | 1.1 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.4 | 1.4 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.3 | 1.7 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.3 | 2.4 | GO:0060022 | hard palate development(GO:0060022) |
| 0.3 | 1.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 0.9 | GO:0097350 | DNA rewinding(GO:0036292) neutrophil clearance(GO:0097350) |
| 0.3 | 0.9 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.3 | 1.6 | GO:0060921 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.3 | 0.8 | GO:0055130 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.3 | 0.8 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.3 | 0.8 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.2 | 3.9 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 1.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 1.1 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 0.5 | GO:0031179 | peptide modification(GO:0031179) |
| 0.2 | 3.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 0.5 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.2 | 2.0 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.2 | 0.8 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.2 | 0.6 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.2 | 0.6 | GO:2000722 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) response to mineralocorticoid(GO:0051385) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.2 | 3.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 4.2 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 2.2 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.2 | 1.4 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.2 | 0.9 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.2 | 0.7 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 1.7 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.2 | 0.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 1.5 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 1.8 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 | 0.5 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.1 | 1.5 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 0.4 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 1.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.4 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.1 | 3.1 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.1 | 1.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.2 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 0.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.5 | GO:0071898 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 | 0.6 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 1.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.5 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.8 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.3 | GO:0071676 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 | 1.9 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.5 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.3 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.1 | 0.4 | GO:2000569 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.1 | 0.6 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 1.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 1.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 1.7 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 1.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 2.6 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.4 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.1 | 0.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.6 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.6 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.2 | GO:2000836 | positive regulation of androgen secretion(GO:2000836) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 1.7 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.2 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 0.3 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.1 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.1 | 0.7 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.8 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.1 | 0.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.3 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 0.2 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.1 | 0.3 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.2 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 1.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 2.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.2 | GO:0002587 | negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.1 | 1.7 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 0.6 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.2 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.1 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.4 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.4 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 1.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.9 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.9 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 2.0 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.6 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 4.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.9 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.5 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 1.5 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.5 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.8 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.7 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.5 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.5 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.8 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.4 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.5 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 2.4 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.6 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.7 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 2.2 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) |
| 0.0 | 0.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 1.0 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.5 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.4 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.0 | 1.1 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 1.0 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.2 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 | 0.4 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 1.7 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.1 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.7 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.1 | GO:0060948 | zygotic specification of dorsal/ventral axis(GO:0007352) cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 1.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.0 | GO:1900150 | antifungal innate immune response(GO:0061760) regulation of defense response to fungus(GO:1900150) regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.7 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.3 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.0 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.8 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.9 | GO:0071806 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.3 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.3 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.0 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.0 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.4 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.7 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.0 | 0.2 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.0 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 2.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.4 | 1.6 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.4 | 1.2 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.4 | 1.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 1.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.3 | 2.0 | GO:0071547 | piP-body(GO:0071547) |
| 0.3 | 1.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 2.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 3.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 1.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 0.5 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.2 | 2.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 2.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 7.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 2.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.3 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 1.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 1.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 3.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.5 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 1.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.4 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.1 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.9 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 1.2 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 1.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 1.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 2.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 2.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 5.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 5.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 2.5 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.4 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 1.1 | 4.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.8 | 5.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.5 | 3.8 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.4 | 1.2 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.3 | 2.5 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.3 | 1.5 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.3 | 1.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.3 | 1.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.3 | 0.8 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 3.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 4.2 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.2 | 0.6 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 3.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 0.6 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.2 | 0.8 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.2 | 0.6 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.2 | 0.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 2.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 5.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 1.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 1.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 0.9 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 2.0 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.7 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 1.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 3.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.5 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 0.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 4.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.4 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 1.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.5 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 0.4 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 2.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.4 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 1.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.3 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 0.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 2.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 2.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 7.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 2.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.5 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.6 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.5 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.6 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 2.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 2.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 0.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 1.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.4 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 4.9 | GO:0070035 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.9 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 0.8 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.5 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.4 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 1.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 3.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.5 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.0 | 0.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 1.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.0 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 1.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.0 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.5 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 4.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.0 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 1.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 4.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 3.3 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 4.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 5.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 5.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 3.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 3.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 5.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 4.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 4.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.8 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 1.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 3.7 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 2.5 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.4 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 2.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 4.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.0 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.0 | 1.5 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.6 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.5 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 1.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME S PHASE | Genes involved in S Phase |