Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Foxd1
Z-value: 0.96
Transcription factors associated with Foxd1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxd1
|
ENSMUSG00000078302.3 | forkhead box D1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxd1 | mm10_v2_chr13_+_98354234_98354250 | 0.20 | 2.4e-01 | Click! |
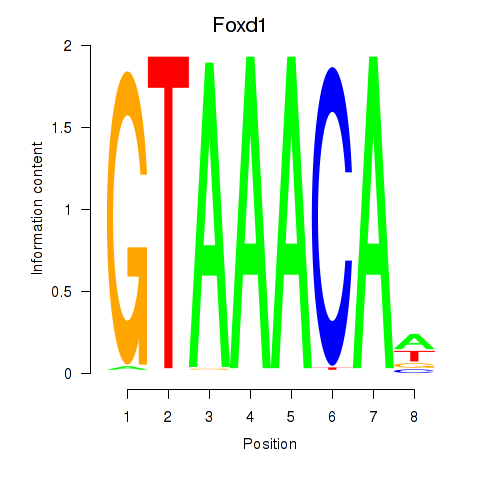
Activity profile of Foxd1 motif
Sorted Z-values of Foxd1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_138170992 | 6.42 |
ENSMUST00000139983.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr11_-_102365111 | 6.11 |
ENSMUST00000006749.9
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr8_+_70373541 | 6.05 |
ENSMUST00000003659.7
|
Comp
|
cartilage oligomeric matrix protein |
| chr5_-_138171248 | 5.87 |
ENSMUST00000153867.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr5_-_138171216 | 5.56 |
ENSMUST00000147920.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr7_+_110772604 | 5.54 |
ENSMUST00000005829.6
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr15_+_80623499 | 5.39 |
ENSMUST00000043149.7
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr11_+_74619594 | 5.26 |
ENSMUST00000100866.2
|
E130309D14Rik
|
RIKEN cDNA E130309D14 gene |
| chr8_-_85380964 | 5.11 |
ENSMUST00000122452.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr10_+_115817247 | 4.33 |
ENSMUST00000035563.7
ENSMUST00000080630.3 ENSMUST00000179196.1 |
Tspan8
|
tetraspanin 8 |
| chr4_+_134315112 | 4.18 |
ENSMUST00000105875.1
ENSMUST00000030638.6 |
Trim63
|
tripartite motif-containing 63 |
| chr6_+_30541582 | 4.16 |
ENSMUST00000096066.4
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr15_+_35296090 | 4.09 |
ENSMUST00000022952.4
|
Osr2
|
odd-skipped related 2 |
| chr2_-_84822546 | 4.08 |
ENSMUST00000028471.5
|
Smtnl1
|
smoothelin-like 1 |
| chr14_+_27000362 | 4.01 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chrX_+_164140447 | 3.85 |
ENSMUST00000073973.4
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr11_+_69095217 | 3.57 |
ENSMUST00000101004.2
|
Per1
|
period circadian clock 1 |
| chr4_+_11191726 | 3.23 |
ENSMUST00000029866.9
ENSMUST00000108324.3 |
Ccne2
|
cyclin E2 |
| chr3_-_52104891 | 3.19 |
ENSMUST00000121440.1
|
Maml3
|
mastermind like 3 (Drosophila) |
| chr19_+_58759700 | 3.17 |
ENSMUST00000026081.3
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr3_-_27153782 | 3.12 |
ENSMUST00000175857.1
ENSMUST00000177055.1 ENSMUST00000176535.1 |
Ect2
|
ect2 oncogene |
| chr9_+_96196246 | 3.07 |
ENSMUST00000165120.2
ENSMUST00000034982.9 |
Tfdp2
|
transcription factor Dp 2 |
| chr6_-_83536215 | 2.91 |
ENSMUST00000075161.5
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr15_-_58324161 | 2.81 |
ENSMUST00000022985.1
|
Klhl38
|
kelch-like 38 |
| chr2_+_14873656 | 2.79 |
ENSMUST00000114718.1
ENSMUST00000114719.1 |
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr3_-_27153844 | 2.79 |
ENSMUST00000176242.2
ENSMUST00000176780.1 |
Ect2
|
ect2 oncogene |
| chr16_-_76022266 | 2.70 |
ENSMUST00000114240.1
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr17_+_47505211 | 2.70 |
ENSMUST00000182935.1
ENSMUST00000182506.1 |
Ccnd3
|
cyclin D3 |
| chr17_+_47505149 | 2.70 |
ENSMUST00000183177.1
ENSMUST00000182848.1 |
Ccnd3
|
cyclin D3 |
| chr6_+_142298419 | 2.67 |
ENSMUST00000041993.2
|
Iapp
|
islet amyloid polypeptide |
| chr17_+_47505043 | 2.66 |
ENSMUST00000182129.1
ENSMUST00000171031.1 |
Ccnd3
|
cyclin D3 |
| chr17_+_47505117 | 2.66 |
ENSMUST00000183044.1
ENSMUST00000037333.10 |
Ccnd3
|
cyclin D3 |
| chr3_-_27153861 | 2.62 |
ENSMUST00000108300.1
ENSMUST00000108298.2 |
Ect2
|
ect2 oncogene |
| chr15_+_79348061 | 2.52 |
ENSMUST00000163691.1
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr17_+_47505073 | 2.51 |
ENSMUST00000183210.1
|
Ccnd3
|
cyclin D3 |
| chr13_-_98815408 | 2.45 |
ENSMUST00000040340.8
ENSMUST00000099277.4 ENSMUST00000179563.1 ENSMUST00000109403.1 |
Fcho2
|
FCH domain only 2 |
| chr15_+_79347534 | 2.34 |
ENSMUST00000096350.3
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr11_+_3330781 | 2.32 |
ENSMUST00000136536.1
ENSMUST00000093399.4 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr3_+_27154020 | 2.22 |
ENSMUST00000181124.1
|
1700125G22Rik
|
RIKEN cDNA 1700125G22 gene |
| chr4_+_11191354 | 2.17 |
ENSMUST00000170901.1
|
Ccne2
|
cyclin E2 |
| chr1_-_45503282 | 2.14 |
ENSMUST00000086430.4
|
Col5a2
|
collagen, type V, alpha 2 |
| chr5_-_149051300 | 2.14 |
ENSMUST00000110505.1
|
Hmgb1
|
high mobility group box 1 |
| chr7_-_115824699 | 2.04 |
ENSMUST00000169129.1
|
Sox6
|
SRY-box containing gene 6 |
| chr14_-_70766598 | 2.01 |
ENSMUST00000167242.1
ENSMUST00000022696.6 |
Xpo7
|
exportin 7 |
| chr12_+_95695350 | 1.88 |
ENSMUST00000110117.1
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr2_+_112265809 | 1.87 |
ENSMUST00000110991.2
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr11_+_96929367 | 1.86 |
ENSMUST00000062172.5
|
Prr15l
|
proline rich 15-like |
| chr11_+_3330401 | 1.84 |
ENSMUST00000045153.4
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr14_+_11227511 | 1.83 |
ENSMUST00000080237.3
|
Rpl21-ps4
|
ribosomal protein L21, pseudogene 4 |
| chr13_-_51701041 | 1.77 |
ENSMUST00000110042.1
|
Gm15440
|
predicted gene 15440 |
| chr2_-_163645125 | 1.77 |
ENSMUST00000017851.3
|
Serinc3
|
serine incorporator 3 |
| chr13_+_104178797 | 1.71 |
ENSMUST00000022225.5
ENSMUST00000069187.5 |
Trim23
|
tripartite motif-containing 23 |
| chrX_-_7740206 | 1.68 |
ENSMUST00000128289.1
|
Ccdc120
|
coiled-coil domain containing 120 |
| chr11_+_101733011 | 1.67 |
ENSMUST00000129741.1
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr1_+_12718496 | 1.66 |
ENSMUST00000088585.3
|
Sulf1
|
sulfatase 1 |
| chr11_+_3332426 | 1.63 |
ENSMUST00000136474.1
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr9_+_78615501 | 1.63 |
ENSMUST00000093812.4
|
Cd109
|
CD109 antigen |
| chr18_-_47333311 | 1.63 |
ENSMUST00000126684.1
ENSMUST00000156422.1 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chrX_+_68678712 | 1.63 |
ENSMUST00000114654.1
ENSMUST00000114655.1 ENSMUST00000114657.2 ENSMUST00000114653.1 |
Fmr1
|
fragile X mental retardation syndrome 1 |
| chr15_+_61985540 | 1.61 |
ENSMUST00000159327.1
ENSMUST00000167731.1 |
Myc
|
myelocytomatosis oncogene |
| chr19_-_41848076 | 1.60 |
ENSMUST00000059231.2
|
Frat2
|
frequently rearranged in advanced T cell lymphomas 2 |
| chr15_+_61985377 | 1.60 |
ENSMUST00000161976.1
ENSMUST00000022971.7 |
Myc
|
myelocytomatosis oncogene |
| chr16_-_74411292 | 1.59 |
ENSMUST00000117200.1
|
Robo2
|
roundabout homolog 2 (Drosophila) |
| chr3_+_138860489 | 1.59 |
ENSMUST00000121826.1
|
Tspan5
|
tetraspanin 5 |
| chr12_+_71016658 | 1.56 |
ENSMUST00000125125.1
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr3_-_93015669 | 1.55 |
ENSMUST00000107301.1
ENSMUST00000029521.4 |
Crct1
|
cysteine-rich C-terminal 1 |
| chr4_-_47010781 | 1.54 |
ENSMUST00000135777.1
|
Gm568
|
predicted gene 568 |
| chr19_+_53329413 | 1.53 |
ENSMUST00000025998.7
|
Mxi1
|
Max interacting protein 1 |
| chr15_-_51991679 | 1.51 |
ENSMUST00000022927.9
|
Rad21
|
RAD21 homolog (S. pombe) |
| chr15_+_97784355 | 1.46 |
ENSMUST00000117892.1
|
Slc48a1
|
solute carrier family 48 (heme transporter), member 1 |
| chr6_-_148946146 | 1.42 |
ENSMUST00000132696.1
|
Fam60a
|
family with sequence similarity 60, member A |
| chr8_-_84197667 | 1.41 |
ENSMUST00000181282.1
|
Gm26887
|
predicted gene, 26887 |
| chr5_-_122988533 | 1.40 |
ENSMUST00000086200.4
ENSMUST00000156474.1 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chrX_+_68678541 | 1.38 |
ENSMUST00000088546.5
|
Fmr1
|
fragile X mental retardation syndrome 1 |
| chr11_+_101732950 | 1.37 |
ENSMUST00000039152.7
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr2_+_52072823 | 1.32 |
ENSMUST00000112693.2
ENSMUST00000069794.5 |
Rif1
|
Rap1 interacting factor 1 homolog (yeast) |
| chrX_+_35888808 | 1.32 |
ENSMUST00000033419.6
|
Dock11
|
dedicator of cytokinesis 11 |
| chr19_-_7341848 | 1.32 |
ENSMUST00000171393.1
|
Mark2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr5_-_149051604 | 1.29 |
ENSMUST00000093196.4
|
Hmgb1
|
high mobility group box 1 |
| chr4_+_138725282 | 1.29 |
ENSMUST00000030530.4
ENSMUST00000124660.1 |
Pla2g2c
|
phospholipase A2, group IIC |
| chr6_+_135362931 | 1.23 |
ENSMUST00000032330.9
|
Emp1
|
epithelial membrane protein 1 |
| chr11_+_96929260 | 1.23 |
ENSMUST00000054311.5
ENSMUST00000107636.3 |
Prr15l
|
proline rich 15-like |
| chr5_+_25246775 | 1.21 |
ENSMUST00000144971.1
|
Galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 |
| chrX_+_42149288 | 1.20 |
ENSMUST00000115073.2
ENSMUST00000115072.1 |
Stag2
|
stromal antigen 2 |
| chr11_+_34047115 | 1.19 |
ENSMUST00000109329.1
ENSMUST00000169878.2 |
Lcp2
|
lymphocyte cytosolic protein 2 |
| chr1_+_134415378 | 1.17 |
ENSMUST00000027727.8
|
Adipor1
|
adiponectin receptor 1 |
| chr15_+_54410755 | 1.17 |
ENSMUST00000036737.3
|
Colec10
|
collectin sub-family member 10 |
| chr15_-_97831460 | 1.17 |
ENSMUST00000079838.7
ENSMUST00000118294.1 |
Hdac7
|
histone deacetylase 7 |
| chr13_+_55445301 | 1.17 |
ENSMUST00000001115.8
ENSMUST00000099482.3 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr19_-_28010995 | 1.14 |
ENSMUST00000172907.1
ENSMUST00000046898.9 |
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr5_-_122989086 | 1.14 |
ENSMUST00000046073.9
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr6_+_52177498 | 1.12 |
ENSMUST00000070587.3
|
5730596B20Rik
|
RIKEN cDNA 5730596B20 gene |
| chr14_-_110755100 | 1.12 |
ENSMUST00000078386.2
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chrX_+_68678624 | 1.07 |
ENSMUST00000114656.1
|
Fmr1
|
fragile X mental retardation syndrome 1 |
| chr2_+_128126030 | 1.06 |
ENSMUST00000089634.5
ENSMUST00000019281.7 ENSMUST00000110341.2 ENSMUST00000103211.1 ENSMUST00000103210.1 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chr5_+_14025305 | 1.06 |
ENSMUST00000073957.6
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr4_+_133130505 | 1.04 |
ENSMUST00000084241.5
ENSMUST00000138831.1 |
Wasf2
|
WAS protein family, member 2 |
| chr19_-_7341792 | 1.04 |
ENSMUST00000164205.1
ENSMUST00000165286.1 ENSMUST00000168324.1 ENSMUST00000032557.8 |
Mark2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr12_-_111980751 | 1.03 |
ENSMUST00000170525.1
|
BC048943
|
cDNA sequence BC048943 |
| chr5_-_122989260 | 1.03 |
ENSMUST00000118027.1
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr11_-_33147400 | 1.03 |
ENSMUST00000020507.7
|
Fgf18
|
fibroblast growth factor 18 |
| chr5_+_31251678 | 1.01 |
ENSMUST00000054829.7
ENSMUST00000114570.1 ENSMUST00000075611.7 |
Krtcap3
|
keratinocyte associated protein 3 |
| chr11_-_120990871 | 1.01 |
ENSMUST00000154483.1
|
Csnk1d
|
casein kinase 1, delta |
| chr2_-_60125651 | 1.01 |
ENSMUST00000112550.1
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr16_-_22439570 | 1.01 |
ENSMUST00000170393.1
|
Etv5
|
ets variant gene 5 |
| chr5_-_147307264 | 0.99 |
ENSMUST00000031650.3
|
Cdx2
|
caudal type homeobox 2 |
| chr2_-_33431324 | 0.99 |
ENSMUST00000113158.1
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr8_+_34807287 | 0.97 |
ENSMUST00000033930.4
|
Dusp4
|
dual specificity phosphatase 4 |
| chr2_-_72813665 | 0.96 |
ENSMUST00000136807.1
ENSMUST00000148327.1 |
6430710C18Rik
|
RIKEN cDNA 6430710C18 gene |
| chr1_+_179546303 | 0.96 |
ENSMUST00000040706.8
|
Cnst
|
consortin, connexin sorting protein |
| chr3_+_134236483 | 0.95 |
ENSMUST00000181904.1
ENSMUST00000053048.9 |
Cxxc4
|
CXXC finger 4 |
| chr1_+_134415414 | 0.95 |
ENSMUST00000112237.1
|
Adipor1
|
adiponectin receptor 1 |
| chr11_-_107348130 | 0.95 |
ENSMUST00000134763.1
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr13_+_75089826 | 0.95 |
ENSMUST00000022075.4
|
Pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr7_+_75455534 | 0.94 |
ENSMUST00000147005.1
ENSMUST00000166315.1 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr5_+_3928033 | 0.93 |
ENSMUST00000143365.1
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr7_+_126776939 | 0.93 |
ENSMUST00000038614.5
ENSMUST00000170882.1 ENSMUST00000106359.1 ENSMUST00000106357.1 ENSMUST00000145762.1 ENSMUST00000132643.1 ENSMUST00000106356.1 |
Ypel3
|
yippee-like 3 (Drosophila) |
| chr6_-_52158292 | 0.92 |
ENSMUST00000000964.5
ENSMUST00000120363.1 |
Hoxa1
|
homeobox A1 |
| chrX_+_101254528 | 0.92 |
ENSMUST00000062000.4
|
Foxo4
|
forkhead box O4 |
| chr2_-_152398046 | 0.92 |
ENSMUST00000063332.8
ENSMUST00000182625.1 |
Sox12
|
SRY-box containing gene 12 |
| chr4_+_101507947 | 0.90 |
ENSMUST00000149047.1
ENSMUST00000106929.3 |
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr8_+_20136455 | 0.90 |
ENSMUST00000179299.1
ENSMUST00000096485.4 |
Gm21811
|
predicted gene, 21811 |
| chr1_-_54926311 | 0.89 |
ENSMUST00000179030.1
ENSMUST00000044359.9 |
Ankrd44
|
ankyrin repeat domain 44 |
| chr17_-_65884902 | 0.86 |
ENSMUST00000024905.9
|
Ralbp1
|
ralA binding protein 1 |
| chr19_-_7341433 | 0.86 |
ENSMUST00000165965.1
ENSMUST00000051711.9 ENSMUST00000169541.1 ENSMUST00000165989.1 |
Mark2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr4_+_132564051 | 0.85 |
ENSMUST00000070690.7
|
Ptafr
|
platelet-activating factor receptor |
| chr7_+_19359740 | 0.84 |
ENSMUST00000140836.1
|
Ppp1r13l
|
protein phosphatase 1, regulatory (inhibitor) subunit 13 like |
| chr7_-_29505447 | 0.84 |
ENSMUST00000183096.1
ENSMUST00000085809.4 |
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr4_+_155791172 | 0.84 |
ENSMUST00000105593.1
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr15_-_96460838 | 0.83 |
ENSMUST00000047835.6
|
Scaf11
|
SR-related CTD-associated factor 11 |
| chr2_-_104028287 | 0.83 |
ENSMUST00000056170.3
|
4931422A03Rik
|
RIKEN cDNA 4931422A03 gene |
| chr12_+_38781093 | 0.83 |
ENSMUST00000161513.1
|
Etv1
|
ets variant gene 1 |
| chr16_+_25286810 | 0.82 |
ENSMUST00000056087.3
|
Tprg
|
transformation related protein 63 regulated |
| chr3_-_102964124 | 0.81 |
ENSMUST00000058899.8
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr4_-_128806045 | 0.81 |
ENSMUST00000106072.2
ENSMUST00000170934.1 |
Zfp362
|
zinc finger protein 362 |
| chr7_-_126776818 | 0.81 |
ENSMUST00000068836.4
|
Gm9967
|
predicted gene 9967 |
| chr4_-_59549243 | 0.81 |
ENSMUST00000173699.1
ENSMUST00000173884.1 ENSMUST00000102883.4 ENSMUST00000174586.1 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr3_+_96557950 | 0.80 |
ENSMUST00000074519.6
ENSMUST00000049093.7 |
Txnip
|
thioredoxin interacting protein |
| chr10_+_53596936 | 0.79 |
ENSMUST00000020004.6
|
Asf1a
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae) |
| chr11_+_120232921 | 0.78 |
ENSMUST00000122148.1
ENSMUST00000044985.7 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr14_-_93888732 | 0.78 |
ENSMUST00000068992.2
|
Pcdh9
|
protocadherin 9 |
| chr2_-_73312701 | 0.77 |
ENSMUST00000058615.9
|
Cir1
|
corepressor interacting with RBPJ, 1 |
| chr13_-_93499803 | 0.76 |
ENSMUST00000065537.7
|
Jmy
|
junction-mediating and regulatory protein |
| chr1_+_55131253 | 0.76 |
ENSMUST00000027122.7
|
Mob4
|
MOB family member 4, phocein |
| chr5_+_21372642 | 0.76 |
ENSMUST00000035799.5
|
Fgl2
|
fibrinogen-like protein 2 |
| chr4_+_136357423 | 0.75 |
ENSMUST00000182167.1
|
Gm17388
|
predicted gene, 17388 |
| chr8_+_19682268 | 0.75 |
ENSMUST00000153710.1
ENSMUST00000127799.1 |
Gm6483
|
predicted gene 6483 |
| chr5_+_107437908 | 0.75 |
ENSMUST00000094541.2
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr18_-_34624562 | 0.73 |
ENSMUST00000003876.3
ENSMUST00000115766.1 ENSMUST00000097626.3 ENSMUST00000115765.1 |
Brd8
|
bromodomain containing 8 |
| chr13_+_118714678 | 0.73 |
ENSMUST00000022246.8
|
Fgf10
|
fibroblast growth factor 10 |
| chr12_-_15816762 | 0.73 |
ENSMUST00000020922.7
|
Trib2
|
tribbles homolog 2 (Drosophila) |
| chr7_-_142372210 | 0.72 |
ENSMUST00000084412.5
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr13_-_78196373 | 0.72 |
ENSMUST00000125176.2
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr5_+_86804508 | 0.72 |
ENSMUST00000038384.7
|
Ythdc1
|
YTH domain containing 1 |
| chr8_-_36953139 | 0.70 |
ENSMUST00000179501.1
|
Dlc1
|
deleted in liver cancer 1 |
| chr1_+_55131317 | 0.70 |
ENSMUST00000162553.1
|
Mob4
|
MOB family member 4, phocein |
| chr11_-_102556122 | 0.69 |
ENSMUST00000143842.1
|
Gpatch8
|
G patch domain containing 8 |
| chr4_+_101507855 | 0.66 |
ENSMUST00000038207.5
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chrX_+_151521146 | 0.66 |
ENSMUST00000112670.1
ENSMUST00000046962.4 ENSMUST00000112668.2 ENSMUST00000046950.6 |
Phf8
|
PHD finger protein 8 |
| chr6_+_29853746 | 0.66 |
ENSMUST00000064872.6
ENSMUST00000152581.1 ENSMUST00000176265.1 ENSMUST00000154079.1 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr5_-_73632421 | 0.65 |
ENSMUST00000087177.2
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr8_+_20567716 | 0.65 |
ENSMUST00000178995.1
|
Gm21092
|
predicted gene, 21092 |
| chr2_+_26973416 | 0.65 |
ENSMUST00000014996.7
ENSMUST00000102891.3 |
Adamts13
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 13 |
| chr3_-_61365951 | 0.65 |
ENSMUST00000066298.2
|
B430305J03Rik
|
RIKEN cDNA B430305J03 gene |
| chrX_+_151520655 | 0.60 |
ENSMUST00000112666.1
ENSMUST00000168501.1 ENSMUST00000112662.2 |
Phf8
|
PHD finger protein 8 |
| chr8_+_65618009 | 0.59 |
ENSMUST00000110258.1
ENSMUST00000110256.1 ENSMUST00000110255.1 |
March1
|
membrane-associated ring finger (C3HC4) 1 |
| chr6_-_5496296 | 0.58 |
ENSMUST00000019721.4
|
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4 |
| chr12_-_100725028 | 0.58 |
ENSMUST00000043599.6
|
Rps6ka5
|
ribosomal protein S6 kinase, polypeptide 5 |
| chr7_-_25788635 | 0.57 |
ENSMUST00000002677.4
ENSMUST00000085948.4 |
Axl
|
AXL receptor tyrosine kinase |
| chr12_+_52699297 | 0.57 |
ENSMUST00000095737.3
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr12_+_38780817 | 0.55 |
ENSMUST00000160856.1
|
Etv1
|
ets variant gene 1 |
| chr19_-_45783512 | 0.55 |
ENSMUST00000026243.3
|
Mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chrX_+_42149534 | 0.53 |
ENSMUST00000127618.1
|
Stag2
|
stromal antigen 2 |
| chr6_-_99096196 | 0.53 |
ENSMUST00000175886.1
|
Foxp1
|
forkhead box P1 |
| chr9_-_108649349 | 0.52 |
ENSMUST00000013338.8
|
Arih2
|
ariadne homolog 2 (Drosophila) |
| chr8_-_35495487 | 0.51 |
ENSMUST00000033927.6
|
Eri1
|
exoribonuclease 1 |
| chr18_-_39489880 | 0.50 |
ENSMUST00000152853.1
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr2_+_122028544 | 0.50 |
ENSMUST00000028668.7
|
Eif3j1
|
eukaryotic translation initiation factor 3, subunit J1 |
| chr1_+_40681659 | 0.49 |
ENSMUST00000027231.7
|
Slc9a2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2 |
| chr6_+_6248659 | 0.49 |
ENSMUST00000181633.1
ENSMUST00000176283.1 ENSMUST00000175814.1 ENSMUST00000181192.1 |
Gm20619
|
predicted gene 20619 |
| chr5_+_86804214 | 0.48 |
ENSMUST00000119339.1
ENSMUST00000120498.1 |
Ythdc1
|
YTH domain containing 1 |
| chr3_+_129532386 | 0.48 |
ENSMUST00000071402.2
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr12_+_69296676 | 0.48 |
ENSMUST00000021362.4
|
Klhdc2
|
kelch domain containing 2 |
| chr2_+_18064645 | 0.48 |
ENSMUST00000114680.2
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr19_-_28011138 | 0.47 |
ENSMUST00000174850.1
|
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr7_+_24907618 | 0.46 |
ENSMUST00000151121.1
|
Arhgef1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr11_-_120086790 | 0.46 |
ENSMUST00000106227.1
ENSMUST00000106229.1 ENSMUST00000180242.1 |
Azi1
|
5-azacytidine induced gene 1 |
| chr19_+_30030589 | 0.45 |
ENSMUST00000112552.1
|
Uhrf2
|
ubiquitin-like, containing PHD and RING finger domains 2 |
| chr7_-_142061021 | 0.44 |
ENSMUST00000084418.2
|
Mob2
|
MOB kinase activator 2 |
| chr11_-_54956047 | 0.41 |
ENSMUST00000155316.1
ENSMUST00000108889.3 ENSMUST00000126703.1 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr1_-_64956731 | 0.39 |
ENSMUST00000123225.1
|
Plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr8_+_31091593 | 0.38 |
ENSMUST00000161713.1
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr15_+_6422240 | 0.37 |
ENSMUST00000163082.1
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr4_+_32238950 | 0.37 |
ENSMUST00000037416.6
|
Bach2
|
BTB and CNC homology 2 |
| chr6_-_39725193 | 0.37 |
ENSMUST00000101497.3
|
Braf
|
Braf transforming gene |
| chr11_+_98741871 | 0.37 |
ENSMUST00000103139.4
|
Thra
|
thyroid hormone receptor alpha |
| chr11_+_43433720 | 0.36 |
ENSMUST00000126128.1
ENSMUST00000151880.1 ENSMUST00000020681.3 |
Slu7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr6_-_88874597 | 0.36 |
ENSMUST00000061262.4
ENSMUST00000140455.1 ENSMUST00000145780.1 |
Podxl2
|
podocalyxin-like 2 |
| chr2_-_71367749 | 0.36 |
ENSMUST00000151937.1
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr7_+_82175156 | 0.35 |
ENSMUST00000180243.1
|
Sh3gl3
|
SH3-domain GRB2-like 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxd1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 17.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.4 | 4.1 | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 1.1 | 3.4 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 1.0 | 3.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 1.0 | 4.1 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 1.0 | 3.8 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.9 | 2.8 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.9 | 5.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.8 | 5.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.8 | 3.2 | GO:0090094 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.7 | 3.6 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.7 | 3.6 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.6 | 5.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.6 | 1.8 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.6 | 13.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.5 | 1.6 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.5 | 2.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.5 | 1.1 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.5 | 1.6 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.5 | 4.7 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.5 | 8.5 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.4 | 1.3 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.4 | 1.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.4 | 1.7 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.4 | 1.9 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.4 | 10.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.4 | 1.8 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 2.7 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.3 | 1.0 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.3 | 0.9 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.3 | 1.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.3 | 1.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.3 | 3.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.3 | 1.6 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.3 | 1.6 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.3 | 1.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 0.7 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.2 | 0.9 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 5.6 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.2 | 3.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 1.5 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.2 | 0.9 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 0.8 | GO:1902943 | regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.2 | 2.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 0.6 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 | 0.2 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.2 | 0.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 3.2 | GO:0019374 | galactolipid metabolic process(GO:0019374) |
| 0.2 | 1.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 5.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 1.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 1.0 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.2 | 1.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.6 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.6 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.1 | 1.4 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.9 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.4 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.4 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.1 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 1.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.6 | GO:0097048 | dendritic cell apoptotic process(GO:0097048) neutrophil clearance(GO:0097350) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.1 | 1.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.8 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.5 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) |
| 0.1 | 2.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.2 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.1 | 4.9 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 1.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 1.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 2.7 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 0.4 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 1.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 1.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 3.2 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 0.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.2 | GO:1903242 | regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 1.0 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.5 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.2 | GO:0060009 | negative regulation of adenylate cyclase activity(GO:0007194) Sertoli cell development(GO:0060009) |
| 0.1 | 0.5 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.1 | 0.4 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 1.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 3.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 1.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 1.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.3 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.8 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.5 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.8 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.4 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 1.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.0 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 0.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.6 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.8 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.6 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 1.1 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 1.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.6 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 1.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.2 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.2 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0008210 | luteinization(GO:0001553) estrogen metabolic process(GO:0008210) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 1.7 | 8.5 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.2 | 17.8 | GO:0042555 | MCM complex(GO:0042555) |
| 1.0 | 4.1 | GO:1902737 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 0.8 | 2.5 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.7 | 2.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 3.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 1.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.3 | 2.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 13.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.5 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.2 | 3.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 1.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 0.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 1.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 4.1 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.5 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 3.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 6.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 2.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 3.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 4.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 4.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 6.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.3 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.0 | 1.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.8 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.1 | 3.4 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.9 | 6.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.7 | 5.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 5.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.6 | 1.8 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.6 | 4.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.6 | 2.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.5 | 3.8 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.5 | 2.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.4 | 17.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.4 | 1.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.4 | 1.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 1.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 1.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 6.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 1.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.3 | 4.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.3 | 13.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 1.8 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 4.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 3.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 3.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 4.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 2.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 5.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 0.5 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.2 | 0.8 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 1.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 2.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 1.8 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.4 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.1 | 0.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 3.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 5.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.7 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.5 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 11.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 5.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 3.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 1.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 1.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 3.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 3.2 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 2.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.9 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 5.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 17.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 16.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 10.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 3.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 2.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 6.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 7.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 17.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 5.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 5.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 13.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 6.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 3.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 3.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 1.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 10.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 2.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 4.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.9 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 1.8 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 1.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 3.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 4.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 4.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.4 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 1.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 4.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |