Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
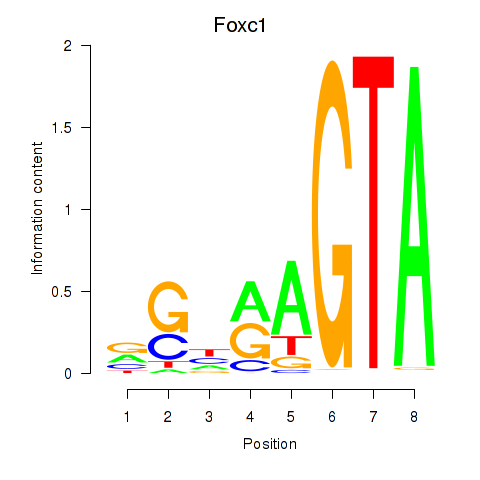
Results for Foxc1
Z-value: 0.67
Transcription factors associated with Foxc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxc1
|
ENSMUSG00000050295.2 | forkhead box C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxc1 | mm10_v2_chr13_+_31806627_31806650 | 0.04 | 8.2e-01 | Click! |
Activity profile of Foxc1 motif
Sorted Z-values of Foxc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_72824482 | 2.75 |
ENSMUST00000047328.4
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr8_+_105048592 | 2.20 |
ENSMUST00000093222.5
ENSMUST00000093223.3 |
Ces3a
|
carboxylesterase 3A |
| chr6_+_68161415 | 2.13 |
ENSMUST00000168090.1
|
Igkv1-115
|
immunoglobulin kappa variable 1-115 |
| chr11_+_70054334 | 2.01 |
ENSMUST00000018699.6
ENSMUST00000108585.2 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr12_+_104101087 | 1.63 |
ENSMUST00000021495.3
|
Serpina5
|
serine (or cysteine) peptidase inhibitor, clade A, member 5 |
| chr4_+_148901128 | 1.52 |
ENSMUST00000147270.1
|
Casz1
|
castor zinc finger 1 |
| chr4_-_32602760 | 1.46 |
ENSMUST00000056517.2
|
Gja10
|
gap junction protein, alpha 10 |
| chr1_-_172895048 | 1.39 |
ENSMUST00000027824.5
|
Apcs
|
serum amyloid P-component |
| chr9_-_103222063 | 1.37 |
ENSMUST00000170904.1
|
Trf
|
transferrin |
| chr9_+_110419750 | 1.27 |
ENSMUST00000035061.6
|
Ngp
|
neutrophilic granule protein |
| chr12_-_113422730 | 1.23 |
ENSMUST00000177715.1
ENSMUST00000103426.1 |
Ighm
|
immunoglobulin heavy constant mu |
| chr3_-_106149761 | 1.20 |
ENSMUST00000149836.1
|
Chi3l3
|
chitinase 3-like 3 |
| chr7_+_142442330 | 1.13 |
ENSMUST00000149529.1
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr15_+_10177623 | 1.06 |
ENSMUST00000124470.1
|
Prlr
|
prolactin receptor |
| chr1_+_136052771 | 0.98 |
ENSMUST00000112068.3
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chrX_+_101449078 | 0.95 |
ENSMUST00000033674.5
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr7_-_44816586 | 0.91 |
ENSMUST00000047356.8
|
Atf5
|
activating transcription factor 5 |
| chr1_+_88166004 | 0.90 |
ENSMUST00000097659.4
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr3_-_123034943 | 0.88 |
ENSMUST00000029761.7
|
Myoz2
|
myozenin 2 |
| chr3_-_15838643 | 0.87 |
ENSMUST00000148194.1
|
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr11_+_108395288 | 0.86 |
ENSMUST00000000049.5
|
Apoh
|
apolipoprotein H |
| chr17_-_31129602 | 0.85 |
ENSMUST00000024827.4
|
Tff3
|
trefoil factor 3, intestinal |
| chr11_+_73160403 | 0.85 |
ENSMUST00000006104.3
|
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr16_-_19883873 | 0.84 |
ENSMUST00000100083.3
|
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chrX_-_95478107 | 0.84 |
ENSMUST00000033549.2
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr6_-_67535783 | 0.79 |
ENSMUST00000058178.4
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr12_-_73047179 | 0.78 |
ENSMUST00000050029.7
|
Six1
|
sine oculis-related homeobox 1 |
| chr4_+_11123950 | 0.76 |
ENSMUST00000142297.1
|
Gm11827
|
predicted gene 11827 |
| chr8_-_11008458 | 0.76 |
ENSMUST00000040514.6
|
Irs2
|
insulin receptor substrate 2 |
| chr14_-_31640878 | 0.76 |
ENSMUST00000167066.1
ENSMUST00000127204.2 ENSMUST00000022437.8 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr11_+_67240557 | 0.74 |
ENSMUST00000170942.1
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle |
| chr7_+_44816364 | 0.74 |
ENSMUST00000118125.1
|
Il4i1
|
interleukin 4 induced 1 |
| chr17_+_48359891 | 0.73 |
ENSMUST00000024792.6
|
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr3_+_68691424 | 0.72 |
ENSMUST00000107816.2
|
Il12a
|
interleukin 12a |
| chr9_+_7558429 | 0.70 |
ENSMUST00000018765.2
|
Mmp8
|
matrix metallopeptidase 8 |
| chr2_+_58754910 | 0.70 |
ENSMUST00000059102.6
|
Upp2
|
uridine phosphorylase 2 |
| chr3_+_95124476 | 0.69 |
ENSMUST00000131597.1
ENSMUST00000005769.6 ENSMUST00000107227.1 |
Tmod4
|
tropomodulin 4 |
| chr2_+_58755177 | 0.68 |
ENSMUST00000102755.3
|
Upp2
|
uridine phosphorylase 2 |
| chr17_+_46254017 | 0.67 |
ENSMUST00000095262.4
|
Lrrc73
|
leucine rich repeat containing 73 |
| chr2_-_104849465 | 0.66 |
ENSMUST00000126824.1
|
Prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr3_+_94377432 | 0.65 |
ENSMUST00000107292.1
|
Rorc
|
RAR-related orphan receptor gamma |
| chr17_+_80944611 | 0.62 |
ENSMUST00000025092.4
|
Tmem178
|
transmembrane protein 178 |
| chr17_+_34238896 | 0.62 |
ENSMUST00000095342.3
|
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr1_-_71653162 | 0.61 |
ENSMUST00000055226.6
|
Fn1
|
fibronectin 1 |
| chr10_+_115569986 | 0.60 |
ENSMUST00000173620.1
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr2_-_155514796 | 0.59 |
ENSMUST00000029131.4
|
Ggt7
|
gamma-glutamyltransferase 7 |
| chr7_-_142656018 | 0.58 |
ENSMUST00000178921.1
|
Igf2
|
insulin-like growth factor 2 |
| chr9_-_45009590 | 0.58 |
ENSMUST00000102832.1
|
Cd3e
|
CD3 antigen, epsilon polypeptide |
| chr18_+_33464163 | 0.57 |
ENSMUST00000097634.3
|
Gm10549
|
predicted gene 10549 |
| chr8_-_69089200 | 0.57 |
ENSMUST00000037478.6
|
Slc18a1
|
solute carrier family 18 (vesicular monoamine), member 1 |
| chr2_-_122298165 | 0.56 |
ENSMUST00000053734.5
|
Duox2
|
dual oxidase 2 |
| chr8_+_105083753 | 0.56 |
ENSMUST00000093221.6
ENSMUST00000074403.6 |
Ces3b
|
carboxylesterase 3B |
| chr3_+_94377505 | 0.56 |
ENSMUST00000098877.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr17_+_34238914 | 0.54 |
ENSMUST00000167280.1
|
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr14_-_37110087 | 0.54 |
ENSMUST00000179488.1
|
2610528A11Rik
|
RIKEN cDNA 2610528A11 gene |
| chr3_+_87435820 | 0.53 |
ENSMUST00000178261.1
ENSMUST00000049926.8 ENSMUST00000166297.1 |
Fcrl5
|
Fc receptor-like 5 |
| chr2_+_130012336 | 0.52 |
ENSMUST00000110299.2
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr2_-_156839790 | 0.52 |
ENSMUST00000134838.1
ENSMUST00000137463.1 ENSMUST00000149275.2 |
Gm14230
|
predicted gene 14230 |
| chrX_+_157698910 | 0.51 |
ENSMUST00000136141.1
|
Smpx
|
small muscle protein, X-linked |
| chr11_-_102296618 | 0.50 |
ENSMUST00000107132.2
ENSMUST00000073234.2 |
Atxn7l3
|
ataxin 7-like 3 |
| chr4_-_94928789 | 0.50 |
ENSMUST00000030309.5
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr1_+_134182404 | 0.49 |
ENSMUST00000153856.1
ENSMUST00000082060.3 ENSMUST00000133701.1 ENSMUST00000132873.1 |
Chi3l1
|
chitinase 3-like 1 |
| chr11_+_61126747 | 0.49 |
ENSMUST00000010286.1
ENSMUST00000146033.1 ENSMUST00000139422.1 |
Tnfrsf13b
|
tumor necrosis factor receptor superfamily, member 13b |
| chr5_-_34187670 | 0.47 |
ENSMUST00000042701.6
ENSMUST00000119171.1 |
Mxd4
|
Max dimerization protein 4 |
| chr8_-_105933832 | 0.47 |
ENSMUST00000034368.6
|
Ctrl
|
chymotrypsin-like |
| chr19_-_40994133 | 0.47 |
ENSMUST00000117695.1
|
Blnk
|
B cell linker |
| chr7_+_30493622 | 0.46 |
ENSMUST00000058280.6
ENSMUST00000133318.1 ENSMUST00000142575.1 ENSMUST00000131040.1 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr3_-_14808358 | 0.45 |
ENSMUST00000181860.1
ENSMUST00000144327.2 |
Car1
|
carbonic anhydrase 1 |
| chr13_+_65512678 | 0.45 |
ENSMUST00000081471.2
|
Gm10139
|
predicted gene 10139 |
| chr4_-_133212480 | 0.44 |
ENSMUST00000052090.8
|
Gpr3
|
G-protein coupled receptor 3 |
| chr16_-_38800193 | 0.43 |
ENSMUST00000057767.4
|
Upk1b
|
uroplakin 1B |
| chr15_-_100599983 | 0.40 |
ENSMUST00000073837.6
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr5_+_114175889 | 0.39 |
ENSMUST00000146841.1
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr14_-_70175397 | 0.39 |
ENSMUST00000143393.1
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr8_+_117701937 | 0.39 |
ENSMUST00000034304.7
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr2_+_172370658 | 0.39 |
ENSMUST00000151511.1
ENSMUST00000116375.1 |
Cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1 |
| chr1_+_107511416 | 0.38 |
ENSMUST00000009356.4
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr11_-_53618659 | 0.38 |
ENSMUST00000000889.6
|
Il4
|
interleukin 4 |
| chr2_-_172370506 | 0.36 |
ENSMUST00000109139.1
ENSMUST00000028997.7 ENSMUST00000109140.3 |
Aurka
|
aurora kinase A |
| chr1_+_107511489 | 0.36 |
ENSMUST00000064916.2
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr17_+_37045980 | 0.36 |
ENSMUST00000174456.1
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr16_-_94370450 | 0.36 |
ENSMUST00000138514.1
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr14_+_31641051 | 0.36 |
ENSMUST00000090147.6
|
Btd
|
biotinidase |
| chr6_+_145121727 | 0.35 |
ENSMUST00000032396.6
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr15_-_58034289 | 0.35 |
ENSMUST00000100655.3
|
9130401M01Rik
|
RIKEN cDNA 9130401M01 gene |
| chr3_+_95160449 | 0.34 |
ENSMUST00000090823.1
ENSMUST00000090821.3 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr11_+_105092194 | 0.33 |
ENSMUST00000021029.5
|
Efcab3
|
EF-hand calcium binding domain 3 |
| chr11_-_84167466 | 0.32 |
ENSMUST00000050771.7
|
Gm11437
|
predicted gene 11437 |
| chr2_+_122315672 | 0.31 |
ENSMUST00000099461.3
|
Duox1
|
dual oxidase 1 |
| chr3_-_95282076 | 0.30 |
ENSMUST00000015855.7
|
Prune
|
prune homolog (Drosophila) |
| chr5_+_134099704 | 0.29 |
ENSMUST00000016088.8
|
Gatsl2
|
GATS protein-like 2 |
| chr6_-_83536215 | 0.29 |
ENSMUST00000075161.5
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr13_+_53525703 | 0.29 |
ENSMUST00000081132.4
|
Gm5449
|
predicted pseudogene 5449 |
| chr18_-_78123324 | 0.29 |
ENSMUST00000160292.1
ENSMUST00000091813.5 |
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr15_-_100599864 | 0.29 |
ENSMUST00000177247.2
ENSMUST00000177505.2 |
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr9_-_51278540 | 0.29 |
ENSMUST00000114427.3
|
Gm684
|
predicted gene 684 |
| chr6_-_55175019 | 0.28 |
ENSMUST00000003569.5
|
Inmt
|
indolethylamine N-methyltransferase |
| chr9_-_60838200 | 0.28 |
ENSMUST00000063858.7
|
Gm9869
|
predicted gene 9869 |
| chr2_+_30807826 | 0.28 |
ENSMUST00000041830.3
ENSMUST00000152374.1 |
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr2_+_151542483 | 0.27 |
ENSMUST00000044011.5
|
Fkbp1a
|
FK506 binding protein 1a |
| chr6_-_129660556 | 0.27 |
ENSMUST00000119533.1
ENSMUST00000145984.1 ENSMUST00000118401.1 ENSMUST00000071920.4 |
Klrc2
|
killer cell lectin-like receptor subfamily C, member 2 |
| chr6_+_48895243 | 0.27 |
ENSMUST00000031835.7
|
Aoc1
|
amine oxidase, copper-containing 1 |
| chr15_+_102503722 | 0.27 |
ENSMUST00000096145.4
|
Gm10337
|
predicted gene 10337 |
| chr8_-_13494479 | 0.26 |
ENSMUST00000033828.5
|
Gas6
|
growth arrest specific 6 |
| chr13_-_34077992 | 0.26 |
ENSMUST00000056427.8
|
Tubb2a
|
tubulin, beta 2A class IIA |
| chr11_+_69935796 | 0.26 |
ENSMUST00000018698.5
|
Ybx2
|
Y box protein 2 |
| chr7_+_129257027 | 0.26 |
ENSMUST00000094018.4
|
Ppapdc1a
|
phosphatidic acid phosphatase type 2 domain containing 1A |
| chr19_+_7268296 | 0.26 |
ENSMUST00000066646.4
|
Rcor2
|
REST corepressor 2 |
| chr4_-_94928820 | 0.25 |
ENSMUST00000107097.2
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr8_-_24576297 | 0.25 |
ENSMUST00000033953.7
ENSMUST00000121992.1 |
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr5_+_138229822 | 0.25 |
ENSMUST00000159798.1
ENSMUST00000159964.1 |
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr13_-_29855630 | 0.25 |
ENSMUST00000091674.5
ENSMUST00000006353.7 |
Cdkal1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr17_-_70851710 | 0.24 |
ENSMUST00000166395.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr9_+_72806874 | 0.23 |
ENSMUST00000055535.8
|
Prtg
|
protogenin homolog (Gallus gallus) |
| chr15_-_76656905 | 0.22 |
ENSMUST00000176274.1
|
Cyhr1
|
cysteine and histidine rich 1 |
| chr3_-_57294880 | 0.22 |
ENSMUST00000171384.1
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr5_-_139484420 | 0.22 |
ENSMUST00000150992.1
|
Zfand2a
|
zinc finger, AN1-type domain 2A |
| chr1_+_45311538 | 0.22 |
ENSMUST00000087883.6
|
Col3a1
|
collagen, type III, alpha 1 |
| chr13_+_42680565 | 0.21 |
ENSMUST00000128646.1
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr12_+_4234023 | 0.21 |
ENSMUST00000179139.1
|
Ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr6_+_55203381 | 0.21 |
ENSMUST00000053094.7
|
Fam188b
|
family with sequence similarity 188, member B |
| chr7_-_44892358 | 0.20 |
ENSMUST00000003049.6
|
Med25
|
mediator of RNA polymerase II transcription, subunit 25 homolog (yeast) |
| chr18_-_33463615 | 0.18 |
ENSMUST00000051087.8
|
Nrep
|
neuronal regeneration related protein |
| chr16_+_18248866 | 0.18 |
ENSMUST00000115640.1
ENSMUST00000140206.1 |
Trmt2a
|
TRM2 tRNA methyltransferase 2A |
| chr17_-_45686214 | 0.18 |
ENSMUST00000113523.2
|
Tmem63b
|
transmembrane protein 63b |
| chr7_-_143502515 | 0.17 |
ENSMUST00000010904.4
|
Phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr1_+_120602405 | 0.17 |
ENSMUST00000079721.7
|
En1
|
engrailed 1 |
| chr9_-_67049143 | 0.17 |
ENSMUST00000113687.1
ENSMUST00000113693.1 ENSMUST00000113701.1 ENSMUST00000034928.5 ENSMUST00000113685.3 ENSMUST00000030185.4 ENSMUST00000050905.9 ENSMUST00000113705.1 ENSMUST00000113697.1 ENSMUST00000113707.2 |
Tpm1
|
tropomyosin 1, alpha |
| chr9_+_122117375 | 0.17 |
ENSMUST00000118886.1
|
Snrk
|
SNF related kinase |
| chr4_+_117550326 | 0.17 |
ENSMUST00000037127.8
|
Eri3
|
exoribonuclease 3 |
| chr5_-_33433976 | 0.16 |
ENSMUST00000173348.1
|
Nkx1-1
|
NK1 transcription factor related, locus 1 (Drosophila) |
| chr15_+_6422240 | 0.16 |
ENSMUST00000163082.1
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr4_+_43267165 | 0.16 |
ENSMUST00000107942.2
ENSMUST00000102953.3 |
Atp8b5
|
ATPase, class I, type 8B, member 5 |
| chr9_-_86880414 | 0.16 |
ENSMUST00000074501.5
ENSMUST00000098495.3 ENSMUST00000074468.6 ENSMUST00000036347.6 |
Snap91
|
synaptosomal-associated protein 91 |
| chr14_-_31128924 | 0.16 |
ENSMUST00000064032.4
ENSMUST00000049732.5 ENSMUST00000090205.3 |
Smim4
|
small itegral membrane protein 4 |
| chr6_+_113472276 | 0.16 |
ENSMUST00000147316.1
|
Il17rc
|
interleukin 17 receptor C |
| chr6_-_29212240 | 0.16 |
ENSMUST00000160878.1
ENSMUST00000078155.5 |
Impdh1
|
inosine 5'-phosphate dehydrogenase 1 |
| chr9_-_83146601 | 0.16 |
ENSMUST00000162246.2
ENSMUST00000161796.2 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr13_-_54766553 | 0.15 |
ENSMUST00000036825.7
|
Sncb
|
synuclein, beta |
| chr17_-_36951338 | 0.15 |
ENSMUST00000173540.1
|
Ppp1r11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr2_-_17390953 | 0.15 |
ENSMUST00000177966.1
|
Nebl
|
nebulette |
| chr2_-_76215363 | 0.14 |
ENSMUST00000144892.1
|
Pde11a
|
phosphodiesterase 11A |
| chr15_+_79108911 | 0.14 |
ENSMUST00000040320.8
|
Micall1
|
microtubule associated monooxygenase, calponin and LIM domain containing -like 1 |
| chr1_+_54250673 | 0.14 |
ENSMUST00000027128.4
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr15_-_102257449 | 0.14 |
ENSMUST00000043172.8
|
Rarg
|
retinoic acid receptor, gamma |
| chr6_+_42405434 | 0.14 |
ENSMUST00000070178.4
|
Tas2r135
|
taste receptor, type 2, member 135 |
| chr10_+_127323700 | 0.14 |
ENSMUST00000069548.5
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr9_-_71592312 | 0.13 |
ENSMUST00000166112.1
|
Myzap
|
myocardial zonula adherens protein |
| chr18_+_78349754 | 0.12 |
ENSMUST00000164064.1
|
Gm6133
|
predicted gene 6133 |
| chr11_+_101082565 | 0.12 |
ENSMUST00000001806.3
ENSMUST00000107308.3 |
Coasy
|
Coenzyme A synthase |
| chr5_+_88487982 | 0.12 |
ENSMUST00000031222.8
|
Enam
|
enamelin |
| chr10_+_77864623 | 0.12 |
ENSMUST00000092366.2
|
Tspear
|
thrombospondin type laminin G domain and EAR repeats |
| chr8_-_25840440 | 0.12 |
ENSMUST00000110609.1
|
Ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr15_+_78597047 | 0.11 |
ENSMUST00000043069.5
|
Cyth4
|
cytohesin 4 |
| chr17_+_16972910 | 0.11 |
ENSMUST00000071374.5
|
BC002059
|
cDNA sequence BC002059 |
| chr5_-_97111589 | 0.10 |
ENSMUST00000069453.2
ENSMUST00000112968.1 |
Paqr3
|
progestin and adipoQ receptor family member III |
| chr11_+_68968107 | 0.10 |
ENSMUST00000102606.3
ENSMUST00000018884.5 |
Slc25a35
|
solute carrier family 25, member 35 |
| chr9_-_96631487 | 0.10 |
ENSMUST00000128346.1
ENSMUST00000034984.6 |
Rasa2
|
RAS p21 protein activator 2 |
| chrX_-_57338598 | 0.10 |
ENSMUST00000033468.4
ENSMUST00000114736.1 |
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr3_-_141982224 | 0.09 |
ENSMUST00000029948.8
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr12_-_85374696 | 0.09 |
ENSMUST00000040766.7
|
Tmed10
|
transmembrane emp24-like trafficking protein 10 (yeast) |
| chr15_-_102257306 | 0.09 |
ENSMUST00000135466.1
|
Rarg
|
retinoic acid receptor, gamma |
| chr16_+_18248961 | 0.09 |
ENSMUST00000100099.3
|
Trmt2a
|
TRM2 tRNA methyltransferase 2A |
| chr2_-_79908389 | 0.09 |
ENSMUST00000090756.4
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr11_-_71201456 | 0.09 |
ENSMUST00000108515.2
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chrX_+_120290259 | 0.08 |
ENSMUST00000113358.3
ENSMUST00000050239.9 ENSMUST00000113364.3 |
Pcdh11x
|
protocadherin 11 X-linked |
| chr6_+_86371489 | 0.08 |
ENSMUST00000089558.5
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr15_+_80623499 | 0.08 |
ENSMUST00000043149.7
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr1_-_131097535 | 0.08 |
ENSMUST00000016672.4
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chr3_+_61364507 | 0.08 |
ENSMUST00000049064.2
|
Rap2b
|
RAP2B, member of RAS oncogene family |
| chr17_-_36951636 | 0.08 |
ENSMUST00000040402.7
ENSMUST00000174711.1 |
Ppp1r11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr7_+_44816088 | 0.08 |
ENSMUST00000057195.9
ENSMUST00000107891.1 |
Nup62
|
nucleoporin 62 |
| chr4_-_141078302 | 0.07 |
ENSMUST00000030760.8
|
Necap2
|
NECAP endocytosis associated 2 |
| chr17_-_45685973 | 0.07 |
ENSMUST00000145873.1
|
Tmem63b
|
transmembrane protein 63b |
| chr2_-_181365306 | 0.07 |
ENSMUST00000108808.1
ENSMUST00000170190.1 ENSMUST00000127988.1 |
Arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr5_-_100038869 | 0.06 |
ENSMUST00000153442.1
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr15_-_97814830 | 0.06 |
ENSMUST00000121514.1
|
Hdac7
|
histone deacetylase 7 |
| chr17_+_55749978 | 0.06 |
ENSMUST00000025004.6
|
Emr4
|
EGF-like module containing, mucin-like, hormone receptor-like sequence 4 |
| chr13_-_49320219 | 0.06 |
ENSMUST00000110086.1
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr15_-_102667749 | 0.06 |
ENSMUST00000075630.3
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) |
| chr14_+_62998063 | 0.06 |
ENSMUST00000100492.3
|
Defb47
|
defensin beta 47 |
| chr2_-_26140468 | 0.06 |
ENSMUST00000133808.1
|
C330006A16Rik
|
RIKEN cDNA C330006A16 gene |
| chr15_+_88819584 | 0.06 |
ENSMUST00000024042.3
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chrX_-_164250368 | 0.06 |
ENSMUST00000112263.1
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr15_-_78405824 | 0.05 |
ENSMUST00000058659.7
|
Tst
|
thiosulfate sulfurtransferase, mitochondrial |
| chr2_+_181365384 | 0.05 |
ENSMUST00000108807.2
|
Zgpat
|
zinc finger, CCCH-type with G patch domain |
| chr5_+_3571664 | 0.04 |
ENSMUST00000008451.5
|
1700109H08Rik
|
RIKEN cDNA 1700109H08 gene |
| chr14_+_77904365 | 0.04 |
ENSMUST00000169978.1
|
Epsti1
|
epithelial stromal interaction 1 (breast) |
| chr11_+_6415443 | 0.04 |
ENSMUST00000132846.1
|
Ppia
|
peptidylprolyl isomerase A |
| chr2_+_30237680 | 0.04 |
ENSMUST00000113654.1
ENSMUST00000095078.2 |
Lrrc8a
|
leucine rich repeat containing 8A |
| chr9_-_71592346 | 0.04 |
ENSMUST00000093823.1
|
Myzap
|
myocardial zonula adherens protein |
| chr4_+_148602527 | 0.03 |
ENSMUST00000105701.2
ENSMUST00000052060.6 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr4_-_58009117 | 0.03 |
ENSMUST00000102897.3
|
Txndc8
|
thioredoxin domain containing 8 |
| chr15_-_82244716 | 0.03 |
ENSMUST00000089155.4
ENSMUST00000089157.3 |
Cenpm
|
centromere protein M |
| chr16_-_97170707 | 0.03 |
ENSMUST00000056102.7
|
Dscam
|
Down syndrome cell adhesion molecule |
| chr11_-_78550777 | 0.03 |
ENSMUST00000103242.4
|
Tmem97
|
transmembrane protein 97 |
| chr1_-_4360256 | 0.03 |
ENSMUST00000027032.4
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr17_-_51810866 | 0.03 |
ENSMUST00000176669.1
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr3_-_96197580 | 0.03 |
ENSMUST00000016087.3
|
Bola1
|
bolA-like 1 (E. coli) |
| chr11_-_45944910 | 0.03 |
ENSMUST00000129820.1
|
Lsm11
|
U7 snRNP-specific Sm-like protein LSM11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0060220 | camera-type eye photoreceptor cell fate commitment(GO:0060220) negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 0.5 | 1.4 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.4 | 1.2 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.3 | 1.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.3 | 1.6 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.3 | 1.2 | GO:0002587 | negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.2 | 0.6 | GO:0071288 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) cellular response to mercury ion(GO:0071288) |
| 0.2 | 1.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 1.4 | GO:0098707 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.2 | 0.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.6 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 1.0 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 | 0.4 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.8 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.7 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.9 | GO:0034196 | blood coagulation, intrinsic pathway(GO:0007597) acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.6 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 1.2 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.1 | 1.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.3 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 2.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.4 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.6 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.9 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.3 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.1 | 0.5 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 0.9 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.8 | GO:0010748 | negative regulation of B cell apoptotic process(GO:0002903) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 0.2 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.1 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 1.5 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.4 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.5 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.6 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) regulation of enamel mineralization(GO:0070173) |
| 0.0 | 1.0 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 0.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.2 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.7 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.3 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.3 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.0 | 0.5 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.6 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 1.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.4 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.4 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.4 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.2 | 1.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 1.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.7 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 1.4 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.4 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 1.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.9 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0098835 | presynaptic endosome(GO:0098830) presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.7 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.8 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 1.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 2.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 1.4 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.2 | 0.7 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.2 | 1.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 1.4 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 0.7 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.2 | 0.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 1.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.6 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.4 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.6 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 0.9 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 1.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 1.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.8 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.3 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 0.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.3 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 2.2 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.0 | 1.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 1.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 3.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 0.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |