Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Ezh2_Atf2_Ikzf1
Z-value: 1.33
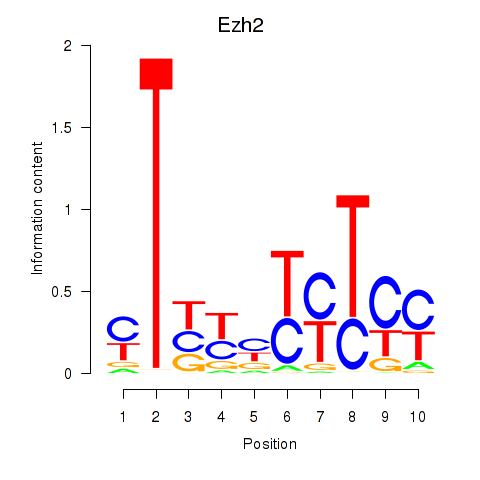
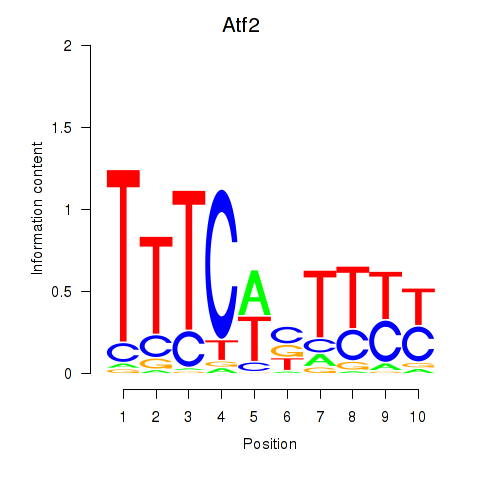
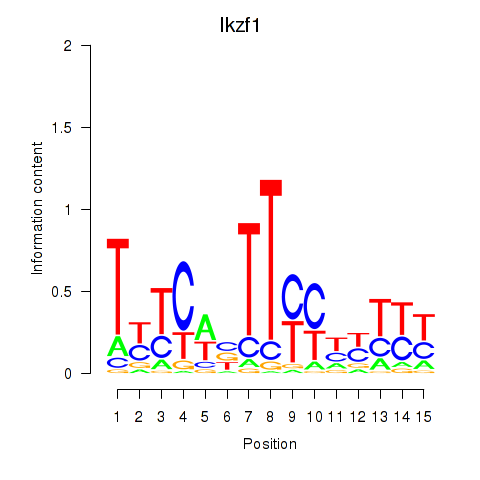
Transcription factors associated with Ezh2_Atf2_Ikzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ezh2
|
ENSMUSG00000029687.10 | enhancer of zeste 2 polycomb repressive complex 2 subunit |
|
Atf2
|
ENSMUSG00000027104.12 | activating transcription factor 2 |
|
Ikzf1
|
ENSMUSG00000018654.11 | IKAROS family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ikzf1 | mm10_v2_chr11_+_11685909_11685947 | -0.60 | 9.7e-05 | Click! |
| Ezh2 | mm10_v2_chr6_-_47594967_47595047 | -0.53 | 7.9e-04 | Click! |
| Atf2 | mm10_v2_chr2_-_73892619_73892639 | 0.04 | 8.2e-01 | Click! |
Activity profile of Ezh2_Atf2_Ikzf1 motif
Sorted Z-values of Ezh2_Atf2_Ikzf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_55175019 | 17.93 |
ENSMUST00000003569.5
|
Inmt
|
indolethylamine N-methyltransferase |
| chr19_-_40073731 | 8.42 |
ENSMUST00000048959.3
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
| chr2_+_67748212 | 6.35 |
ENSMUST00000180887.1
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr6_+_34746368 | 6.14 |
ENSMUST00000142716.1
|
Cald1
|
caldesmon 1 |
| chr19_+_39287074 | 6.09 |
ENSMUST00000003137.8
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr6_+_125321205 | 5.57 |
ENSMUST00000176365.1
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr5_-_92328068 | 4.98 |
ENSMUST00000113093.3
|
Cxcl9
|
chemokine (C-X-C motif) ligand 9 |
| chr19_+_39007019 | 4.83 |
ENSMUST00000025966.4
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr6_+_34745952 | 4.72 |
ENSMUST00000123823.1
ENSMUST00000136907.1 ENSMUST00000126181.1 |
Cald1
|
caldesmon 1 |
| chr15_+_10215955 | 4.58 |
ENSMUST00000130720.1
|
Prlr
|
prolactin receptor |
| chr6_+_125321409 | 4.45 |
ENSMUST00000176442.1
ENSMUST00000177329.1 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr19_+_40089688 | 4.28 |
ENSMUST00000068094.6
ENSMUST00000080171.2 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr10_-_109010955 | 4.02 |
ENSMUST00000105276.1
ENSMUST00000064054.7 |
Syt1
|
synaptotagmin I |
| chr6_-_124542281 | 3.47 |
ENSMUST00000159463.1
ENSMUST00000162844.1 ENSMUST00000160505.1 ENSMUST00000162443.1 |
C1s
|
complement component 1, s subcomponent |
| chr13_+_4434306 | 3.36 |
ENSMUST00000021630.8
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr6_+_41302265 | 3.22 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr1_-_169747634 | 3.15 |
ENSMUST00000027991.5
ENSMUST00000111357.1 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr2_+_173153048 | 3.08 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr16_-_23520579 | 3.08 |
ENSMUST00000089883.5
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr1_+_167618246 | 3.04 |
ENSMUST00000111380.1
|
Rxrg
|
retinoid X receptor gamma |
| chr7_+_100006404 | 2.98 |
ENSMUST00000032977.4
|
Chrdl2
|
chordin-like 2 |
| chr6_+_41521782 | 2.93 |
ENSMUST00000070380.4
|
Prss2
|
protease, serine, 2 |
| chr14_+_28504736 | 2.84 |
ENSMUST00000063465.4
|
Wnt5a
|
wingless-related MMTV integration site 5A |
| chr1_-_193264006 | 2.82 |
ENSMUST00000161737.1
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr1_-_150466165 | 2.78 |
ENSMUST00000162367.1
ENSMUST00000161611.1 ENSMUST00000161320.1 ENSMUST00000159035.1 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr15_+_10314102 | 2.74 |
ENSMUST00000127467.1
|
Prlr
|
prolactin receptor |
| chr6_-_41314700 | 2.73 |
ENSMUST00000064324.5
|
Try5
|
trypsin 5 |
| chr7_+_26835305 | 2.72 |
ENSMUST00000005685.8
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr7_+_25897620 | 2.70 |
ENSMUST00000072438.6
ENSMUST00000005477.5 |
Cyp2b10
|
cytochrome P450, family 2, subfamily b, polypeptide 10 |
| chrX_+_107255878 | 2.69 |
ENSMUST00000101294.2
ENSMUST00000118820.1 ENSMUST00000120971.1 |
Gpr174
|
G protein-coupled receptor 174 |
| chr6_-_114921778 | 2.60 |
ENSMUST00000032459.7
|
Vgll4
|
vestigial like 4 (Drosophila) |
| chr8_-_93079965 | 2.59 |
ENSMUST00000109582.1
|
Ces1b
|
carboxylesterase 1B |
| chr1_-_190170671 | 2.58 |
ENSMUST00000175916.1
|
Prox1
|
prospero-related homeobox 1 |
| chr16_+_43363855 | 2.57 |
ENSMUST00000156367.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr6_-_31218421 | 2.55 |
ENSMUST00000115107.1
|
AB041803
|
cDNA sequence AB041803 |
| chr4_-_82505707 | 2.54 |
ENSMUST00000107248.1
ENSMUST00000107247.1 |
Nfib
|
nuclear factor I/B |
| chr13_+_40917626 | 2.49 |
ENSMUST00000067778.6
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr9_+_46269069 | 2.49 |
ENSMUST00000034584.3
|
Apoa5
|
apolipoprotein A-V |
| chr3_+_97628804 | 2.48 |
ENSMUST00000107050.1
ENSMUST00000029729.8 ENSMUST00000107049.1 |
Fmo5
|
flavin containing monooxygenase 5 |
| chr15_-_5063741 | 2.48 |
ENSMUST00000110689.3
|
C7
|
complement component 7 |
| chr9_-_117252111 | 2.45 |
ENSMUST00000111772.3
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr1_-_190170178 | 2.45 |
ENSMUST00000177288.1
|
Prox1
|
prospero-related homeobox 1 |
| chr10_+_127801145 | 2.41 |
ENSMUST00000071646.1
|
Rdh16
|
retinol dehydrogenase 16 |
| chr18_-_74961252 | 2.39 |
ENSMUST00000066532.4
|
Lipg
|
lipase, endothelial |
| chr19_+_26623419 | 2.37 |
ENSMUST00000176584.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_+_90460889 | 2.36 |
ENSMUST00000031314.8
|
Alb
|
albumin |
| chr16_+_41532851 | 2.30 |
ENSMUST00000078873.4
|
Lsamp
|
limbic system-associated membrane protein |
| chr6_-_144209471 | 2.26 |
ENSMUST00000038815.7
|
Sox5
|
SRY-box containing gene 5 |
| chr5_+_92392585 | 2.23 |
ENSMUST00000126281.1
|
Art3
|
ADP-ribosyltransferase 3 |
| chr2_-_164857542 | 2.17 |
ENSMUST00000109316.1
ENSMUST00000156255.1 ENSMUST00000128110.1 ENSMUST00000109317.3 |
Pltp
|
phospholipid transfer protein |
| chr2_+_93642307 | 2.13 |
ENSMUST00000042078.3
ENSMUST00000111254.1 |
Alx4
|
aristaless-like homeobox 4 |
| chr11_-_69805617 | 2.13 |
ENSMUST00000051025.4
|
Tmem102
|
transmembrane protein 102 |
| chr9_-_117251801 | 2.13 |
ENSMUST00000172564.1
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr10_+_116301374 | 2.08 |
ENSMUST00000092167.5
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr16_-_33056174 | 2.06 |
ENSMUST00000115100.1
ENSMUST00000040309.8 |
Iqcg
|
IQ motif containing G |
| chr4_-_84546284 | 2.04 |
ENSMUST00000177040.1
|
Bnc2
|
basonuclin 2 |
| chr6_-_136875794 | 2.02 |
ENSMUST00000032342.1
|
Mgp
|
matrix Gla protein |
| chr7_+_101378183 | 2.01 |
ENSMUST00000084895.5
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr17_-_78985428 | 2.00 |
ENSMUST00000118991.1
|
Prkd3
|
protein kinase D3 |
| chr10_-_44004846 | 1.99 |
ENSMUST00000020017.8
|
Aim1
|
absent in melanoma 1 |
| chr6_-_28134545 | 1.99 |
ENSMUST00000115323.1
|
Grm8
|
glutamate receptor, metabotropic 8 |
| chr2_-_24048857 | 1.97 |
ENSMUST00000114497.1
|
Hnmt
|
histamine N-methyltransferase |
| chr3_+_19985612 | 1.95 |
ENSMUST00000172860.1
|
Cp
|
ceruloplasmin |
| chr13_+_23870259 | 1.93 |
ENSMUST00000110413.1
|
Slc17a1
|
solute carrier family 17 (sodium phosphate), member 1 |
| chr10_-_109009055 | 1.88 |
ENSMUST00000156979.1
|
Syt1
|
synaptotagmin I |
| chr16_+_43247278 | 1.88 |
ENSMUST00000114691.1
ENSMUST00000079441.6 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr6_+_41392356 | 1.87 |
ENSMUST00000049079.7
|
Gm5771
|
predicted gene 5771 |
| chr16_+_45093611 | 1.87 |
ENSMUST00000099498.2
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr10_+_87861309 | 1.87 |
ENSMUST00000122100.1
|
Igf1
|
insulin-like growth factor 1 |
| chr10_+_57784859 | 1.85 |
ENSMUST00000020024.5
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr4_+_102421518 | 1.84 |
ENSMUST00000106904.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr6_+_121343052 | 1.82 |
ENSMUST00000166457.1
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr10_-_102490418 | 1.81 |
ENSMUST00000020040.3
|
Nts
|
neurotensin |
| chr17_-_90455872 | 1.80 |
ENSMUST00000174337.1
ENSMUST00000172466.1 |
Nrxn1
|
neurexin I |
| chr2_-_173218879 | 1.78 |
ENSMUST00000109116.2
ENSMUST00000029018.7 |
Zbp1
|
Z-DNA binding protein 1 |
| chr2_-_148045891 | 1.77 |
ENSMUST00000109964.1
|
Foxa2
|
forkhead box A2 |
| chr6_-_144209448 | 1.77 |
ENSMUST00000077160.5
|
Sox5
|
SRY-box containing gene 5 |
| chr8_-_3878549 | 1.74 |
ENSMUST00000011445.6
|
Cd209d
|
CD209d antigen |
| chr1_-_174921813 | 1.71 |
ENSMUST00000055294.3
|
Grem2
|
gremlin 2 homolog, cysteine knot superfamily (Xenopus laevis) |
| chr9_-_91365756 | 1.71 |
ENSMUST00000034927.6
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr10_-_95415283 | 1.71 |
ENSMUST00000119917.1
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr5_-_77115145 | 1.70 |
ENSMUST00000081964.5
|
Hopx
|
HOP homeobox |
| chr3_+_19957088 | 1.70 |
ENSMUST00000108328.1
|
Cp
|
ceruloplasmin |
| chr15_+_66891477 | 1.68 |
ENSMUST00000118823.1
|
Wisp1
|
WNT1 inducible signaling pathway protein 1 |
| chr19_+_8664005 | 1.68 |
ENSMUST00000035444.3
ENSMUST00000163785.1 |
Chrm1
|
cholinergic receptor, muscarinic 1, CNS |
| chr6_-_21852509 | 1.68 |
ENSMUST00000031678.3
|
Tspan12
|
tetraspanin 12 |
| chr19_-_58455161 | 1.68 |
ENSMUST00000135730.1
ENSMUST00000152507.1 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chrX_-_72656135 | 1.68 |
ENSMUST00000055966.6
|
Gabra3
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 |
| chr18_+_20310738 | 1.67 |
ENSMUST00000077146.3
|
Dsg1a
|
desmoglein 1 alpha |
| chr3_+_62338344 | 1.66 |
ENSMUST00000079300.6
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr6_+_124304646 | 1.64 |
ENSMUST00000112541.2
ENSMUST00000032234.2 |
Cd163
|
CD163 antigen |
| chr10_+_84756055 | 1.64 |
ENSMUST00000060397.6
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr10_+_57784914 | 1.64 |
ENSMUST00000165013.1
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr7_-_44375006 | 1.64 |
ENSMUST00000107933.1
|
1700008O03Rik
|
RIKEN cDNA 1700008O03 gene |
| chr6_+_125321333 | 1.64 |
ENSMUST00000081440.7
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr10_+_60106198 | 1.63 |
ENSMUST00000121820.2
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr16_+_45094036 | 1.61 |
ENSMUST00000061050.5
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr6_+_121343385 | 1.60 |
ENSMUST00000168295.1
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr13_+_42709482 | 1.60 |
ENSMUST00000066928.5
ENSMUST00000148891.1 |
Phactr1
|
phosphatase and actin regulator 1 |
| chr5_-_87535113 | 1.59 |
ENSMUST00000120150.1
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr16_+_48842552 | 1.58 |
ENSMUST00000023329.4
|
Retnla
|
resistin like alpha |
| chr7_+_140763739 | 1.57 |
ENSMUST00000026552.7
|
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr7_-_80401707 | 1.56 |
ENSMUST00000120753.1
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr10_-_95415484 | 1.56 |
ENSMUST00000172070.1
ENSMUST00000150432.1 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr4_-_49549523 | 1.55 |
ENSMUST00000029987.9
|
Aldob
|
aldolase B, fructose-bisphosphate |
| chr9_-_106476590 | 1.55 |
ENSMUST00000112479.2
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr18_-_61536522 | 1.54 |
ENSMUST00000171629.1
|
Arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr19_-_42202150 | 1.52 |
ENSMUST00000018966.7
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr3_-_113574242 | 1.52 |
ENSMUST00000142505.2
|
Amy1
|
amylase 1, salivary |
| chr16_+_56477838 | 1.51 |
ENSMUST00000048471.7
ENSMUST00000096013.3 ENSMUST00000096012.3 ENSMUST00000171000.1 |
Abi3bp
|
ABI gene family, member 3 (NESH) binding protein |
| chr8_-_24576297 | 1.51 |
ENSMUST00000033953.7
ENSMUST00000121992.1 |
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr19_+_39992424 | 1.49 |
ENSMUST00000049178.2
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr9_-_50555170 | 1.48 |
ENSMUST00000119103.1
|
Bco2
|
beta-carotene oxygenase 2 |
| chr17_+_34204080 | 1.48 |
ENSMUST00000138491.1
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_-_21370452 | 1.47 |
ENSMUST00000102875.4
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr7_-_31054815 | 1.45 |
ENSMUST00000071697.4
ENSMUST00000108110.3 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr9_-_29963112 | 1.45 |
ENSMUST00000075069.4
|
Ntm
|
neurotrimin |
| chr1_+_74713551 | 1.44 |
ENSMUST00000027356.5
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr13_-_115101909 | 1.44 |
ENSMUST00000061673.7
|
Itga1
|
integrin alpha 1 |
| chr19_-_20727533 | 1.44 |
ENSMUST00000025656.3
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr3_+_137341103 | 1.43 |
ENSMUST00000119475.1
|
Emcn
|
endomucin |
| chr15_+_10177623 | 1.43 |
ENSMUST00000124470.1
|
Prlr
|
prolactin receptor |
| chr7_-_126585775 | 1.43 |
ENSMUST00000084589.4
|
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr5_+_92387673 | 1.42 |
ENSMUST00000145072.1
|
Art3
|
ADP-ribosyltransferase 3 |
| chr3_+_19957240 | 1.41 |
ENSMUST00000108325.2
|
Cp
|
ceruloplasmin |
| chr7_+_27119909 | 1.41 |
ENSMUST00000003100.8
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr9_-_106476104 | 1.39 |
ENSMUST00000156426.1
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr6_-_144209558 | 1.39 |
ENSMUST00000111749.1
ENSMUST00000170367.2 |
Sox5
|
SRY-box containing gene 5 |
| chr18_+_20247340 | 1.39 |
ENSMUST00000054128.6
|
Dsg1c
|
desmoglein 1 gamma |
| chr14_+_55561060 | 1.39 |
ENSMUST00000117701.1
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr2_-_67194695 | 1.38 |
ENSMUST00000147939.1
|
Gm13598
|
predicted gene 13598 |
| chr4_+_99030946 | 1.38 |
ENSMUST00000030280.6
|
Angptl3
|
angiopoietin-like 3 |
| chr10_-_20548361 | 1.38 |
ENSMUST00000164195.1
|
Pde7b
|
phosphodiesterase 7B |
| chr3_+_66981352 | 1.37 |
ENSMUST00000162036.1
|
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr16_+_43503607 | 1.37 |
ENSMUST00000126100.1
ENSMUST00000123047.1 ENSMUST00000156981.1 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr2_+_43555321 | 1.36 |
ENSMUST00000028223.2
|
Kynu
|
kynureninase (L-kynurenine hydrolase) |
| chr15_+_22549022 | 1.35 |
ENSMUST00000163361.1
|
Cdh18
|
cadherin 18 |
| chr9_-_106476372 | 1.34 |
ENSMUST00000123555.1
ENSMUST00000125850.1 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr10_+_60106452 | 1.33 |
ENSMUST00000165024.2
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr14_+_52824340 | 1.33 |
ENSMUST00000103648.2
|
Trav11d
|
T cell receptor alpha variable 11D |
| chr6_-_59024340 | 1.33 |
ENSMUST00000173193.1
|
Fam13a
|
family with sequence similarity 13, member A |
| chr5_+_114923234 | 1.32 |
ENSMUST00000031540.4
ENSMUST00000112143.3 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr1_-_155146755 | 1.32 |
ENSMUST00000027744.8
|
Mr1
|
major histocompatibility complex, class I-related |
| chr11_+_61653259 | 1.31 |
ENSMUST00000004959.2
|
Grap
|
GRB2-related adaptor protein |
| chr9_+_43259879 | 1.31 |
ENSMUST00000179013.1
|
D630033O11Rik
|
RIKEN cDNA D630033O11 gene |
| chr1_+_169655493 | 1.31 |
ENSMUST00000027997.3
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr10_+_4611971 | 1.31 |
ENSMUST00000105590.1
ENSMUST00000067086.7 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr2_+_43555342 | 1.31 |
ENSMUST00000112826.1
ENSMUST00000050511.6 |
Kynu
|
kynureninase (L-kynurenine hydrolase) |
| chr10_-_20548320 | 1.31 |
ENSMUST00000169404.1
|
Pde7b
|
phosphodiesterase 7B |
| chr2_+_169633517 | 1.30 |
ENSMUST00000109157.1
|
Tshz2
|
teashirt zinc finger family member 2 |
| chr17_-_84682932 | 1.30 |
ENSMUST00000066175.3
|
Abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chrX_-_162565514 | 1.29 |
ENSMUST00000154424.1
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr11_+_118428493 | 1.29 |
ENSMUST00000017590.2
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr9_-_50693799 | 1.28 |
ENSMUST00000120622.1
|
Dixdc1
|
DIX domain containing 1 |
| chr10_+_87859062 | 1.27 |
ENSMUST00000095360.4
|
Igf1
|
insulin-like growth factor 1 |
| chr12_-_84450944 | 1.26 |
ENSMUST00000085192.5
|
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr19_+_39510844 | 1.26 |
ENSMUST00000025968.4
|
Cyp2c39
|
cytochrome P450, family 2, subfamily c, polypeptide 39 |
| chr11_+_68968107 | 1.26 |
ENSMUST00000102606.3
ENSMUST00000018884.5 |
Slc25a35
|
solute carrier family 25, member 35 |
| chr17_-_34000257 | 1.25 |
ENSMUST00000087189.6
ENSMUST00000173075.1 ENSMUST00000172760.1 ENSMUST00000172912.1 ENSMUST00000025181.10 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr11_-_53773187 | 1.25 |
ENSMUST00000170390.1
|
Gm17334
|
predicted gene, 17334 |
| chr12_-_31559969 | 1.23 |
ENSMUST00000001253.7
|
Slc26a4
|
solute carrier family 26, member 4 |
| chr17_+_84683113 | 1.23 |
ENSMUST00000045714.8
|
Abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr15_+_66891320 | 1.23 |
ENSMUST00000005255.2
|
Wisp1
|
WNT1 inducible signaling pathway protein 1 |
| chr9_-_117252126 | 1.22 |
ENSMUST00000174868.1
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr7_-_48843663 | 1.22 |
ENSMUST00000167786.2
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr2_-_29253001 | 1.22 |
ENSMUST00000071201.4
|
Ntng2
|
netrin G2 |
| chr2_-_28563362 | 1.21 |
ENSMUST00000028161.5
|
Cel
|
carboxyl ester lipase |
| chr3_+_19957037 | 1.21 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr16_+_44173271 | 1.20 |
ENSMUST00000088356.4
ENSMUST00000169582.1 |
Gm608
|
predicted gene 608 |
| chr8_+_46010596 | 1.20 |
ENSMUST00000110381.2
|
Lrp2bp
|
Lrp2 binding protein |
| chr18_-_39489157 | 1.20 |
ENSMUST00000131885.1
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr18_-_3281712 | 1.20 |
ENSMUST00000182204.1
ENSMUST00000154705.1 ENSMUST00000182833.1 ENSMUST00000151084.1 |
Crem
|
cAMP responsive element modulator |
| chr14_-_55884223 | 1.18 |
ENSMUST00000172378.1
|
Cbln3
|
cerebellin 3 precursor protein |
| chr2_+_3713449 | 1.18 |
ENSMUST00000027965.4
|
Fam107b
|
family with sequence similarity 107, member B |
| chr14_+_52810934 | 1.17 |
ENSMUST00000103646.3
|
Trav10d
|
T cell receptor alpha variable 10D |
| chr6_-_138073196 | 1.17 |
ENSMUST00000050132.3
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr4_+_11704439 | 1.16 |
ENSMUST00000108304.2
|
Gem
|
GTP binding protein (gene overexpressed in skeletal muscle) |
| chr2_+_97467657 | 1.15 |
ENSMUST00000059049.7
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr2_+_58755177 | 1.15 |
ENSMUST00000102755.3
|
Upp2
|
uridine phosphorylase 2 |
| chr8_-_84773381 | 1.15 |
ENSMUST00000109764.1
|
Nfix
|
nuclear factor I/X |
| chr14_+_69171576 | 1.14 |
ENSMUST00000062437.8
|
Nkx2-6
|
NK2 homeobox 6 |
| chr14_-_124677089 | 1.13 |
ENSMUST00000095529.3
|
Fgf14
|
fibroblast growth factor 14 |
| chr11_+_60777525 | 1.13 |
ENSMUST00000056907.6
ENSMUST00000102667.3 |
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 homolog (human) |
| chr4_+_43641262 | 1.12 |
ENSMUST00000123351.1
ENSMUST00000128549.1 |
Npr2
|
natriuretic peptide receptor 2 |
| chr10_+_127866457 | 1.12 |
ENSMUST00000092058.3
|
BC089597
|
cDNA sequence BC089597 |
| chr15_-_77533312 | 1.10 |
ENSMUST00000062562.5
|
Apol7c
|
apolipoprotein L 7c |
| chrX_-_70365052 | 1.10 |
ENSMUST00000101509.2
|
Ids
|
iduronate 2-sulfatase |
| chr18_-_38929148 | 1.09 |
ENSMUST00000134864.1
|
Fgf1
|
fibroblast growth factor 1 |
| chr17_+_84683131 | 1.09 |
ENSMUST00000171915.1
|
Abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr1_-_162898665 | 1.09 |
ENSMUST00000111510.1
ENSMUST00000045902.6 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr2_+_58754910 | 1.09 |
ENSMUST00000059102.6
|
Upp2
|
uridine phosphorylase 2 |
| chr6_+_21986887 | 1.09 |
ENSMUST00000151315.1
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr17_+_43389436 | 1.08 |
ENSMUST00000113599.1
|
Gpr116
|
G protein-coupled receptor 116 |
| chr15_+_100304782 | 1.08 |
ENSMUST00000067752.3
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr18_+_37489465 | 1.08 |
ENSMUST00000055949.2
|
Pcdhb18
|
protocadherin beta 18 |
| chr18_+_33464163 | 1.07 |
ENSMUST00000097634.3
|
Gm10549
|
predicted gene 10549 |
| chr5_+_130448801 | 1.07 |
ENSMUST00000111288.2
|
Caln1
|
calneuron 1 |
| chr1_-_20617992 | 1.05 |
ENSMUST00000088448.5
|
Pkhd1
|
polycystic kidney and hepatic disease 1 |
| chr16_+_43364145 | 1.05 |
ENSMUST00000148775.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chrX_-_95166307 | 1.04 |
ENSMUST00000113873.2
ENSMUST00000113876.2 ENSMUST00000113885.1 ENSMUST00000113883.1 ENSMUST00000182001.1 ENSMUST00000113882.1 ENSMUST00000113878.1 ENSMUST00000182562.1 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ezh2_Atf2_Ikzf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0002194 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.6 | 17.5 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 1.5 | 8.8 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 1.4 | 4.3 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 1.3 | 3.8 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.2 | 3.6 | GO:0045796 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 1.1 | 3.4 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.0 | 4.2 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 1.0 | 3.1 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 1.0 | 4.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.9 | 2.8 | GO:0061348 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.9 | 2.7 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.8 | 3.2 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.8 | 2.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.7 | 2.8 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.7 | 2.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.7 | 6.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.6 | 2.6 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.6 | 1.9 | GO:0060197 | cloacal septation(GO:0060197) |
| 0.6 | 17.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.6 | 2.4 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.6 | 1.8 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.6 | 0.6 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.6 | 2.9 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.6 | 5.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.5 | 2.7 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.5 | 5.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.5 | 3.1 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.5 | 2.6 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.5 | 0.5 | GO:1902861 | plasma membrane copper ion transport(GO:0015679) copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.5 | 1.5 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.5 | 1.5 | GO:0019255 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.5 | 1.4 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.5 | 3.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.5 | 1.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.4 | 3.9 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.4 | 1.3 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.4 | 2.9 | GO:0052805 | imidazole-containing compound catabolic process(GO:0052805) |
| 0.4 | 1.2 | GO:1903918 | regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) |
| 0.4 | 1.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.4 | 4.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 2.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 1.6 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.4 | 1.6 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.4 | 1.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.3 | 2.8 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.3 | 2.4 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 2.4 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.3 | 1.0 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.3 | 1.6 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.3 | 1.0 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.3 | 2.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.3 | 3.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 1.5 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.3 | 3.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.3 | 1.4 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.3 | 0.8 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 | 0.8 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of hypersensitivity(GO:0002884) |
| 0.3 | 1.1 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.3 | 0.5 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 1.1 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.3 | 1.6 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.3 | 2.9 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.3 | 1.0 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.3 | 5.7 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.3 | 0.8 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.3 | 0.5 | GO:1904057 | negative regulation of sensory perception of pain(GO:1904057) |
| 0.2 | 1.2 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.2 | 0.5 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.2 | 10.9 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.2 | 1.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 0.7 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.2 | 0.7 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.2 | 0.5 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.2 | 3.6 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.2 | 0.7 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 | 0.4 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 | 6.9 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.2 | 0.6 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 | 0.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.2 | 1.7 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.2 | 0.8 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.2 | 2.5 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.2 | 0.8 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.2 | 0.8 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.2 | 1.0 | GO:0003166 | bundle of His development(GO:0003166) |
| 0.2 | 2.0 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 1.6 | GO:0070189 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.2 | 0.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 1.6 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 0.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.2 | 0.6 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.2 | 0.4 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.2 | 1.0 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.2 | 0.6 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.2 | 1.7 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 0.6 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 1.5 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.2 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 1.3 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.2 | 0.5 | GO:0060618 | nipple development(GO:0060618) |
| 0.2 | 0.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 0.5 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.2 | 2.4 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.2 | 0.9 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.2 | 0.5 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.2 | 0.3 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.2 | 0.5 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 0.2 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.2 | 1.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 | 0.5 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 | 0.2 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.2 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 0.9 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.2 | 0.5 | GO:1904882 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.2 | 0.5 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 1.6 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 2.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.3 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 1.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.2 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 7.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 0.6 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 1.8 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.6 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.1 | 0.9 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.4 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 1.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 2.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.3 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.8 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 2.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.7 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.1 | 0.4 | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.1 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 3.3 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 1.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 2.7 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 0.3 | GO:0072198 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) |
| 0.1 | 0.6 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.1 | 0.3 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 1.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.6 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.3 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.5 | GO:1901341 | positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.8 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 1.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.2 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.1 | 0.7 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.6 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.2 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.1 | 0.6 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.6 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 0.4 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 1.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.5 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.5 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 0.7 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.7 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 | 0.9 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 | 0.3 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 1.0 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.1 | 0.9 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 | 1.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.3 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.9 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.5 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 0.3 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.3 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.1 | 0.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.2 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.3 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.1 | 0.4 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 7.7 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 1.1 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.9 | GO:1904417 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.1 | 0.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 1.5 | GO:2001212 | regulation of vasculogenesis(GO:2001212) |
| 0.1 | 0.2 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 0.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 2.6 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 4.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.1 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 0.1 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.6 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.3 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 1.8 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 0.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 0.8 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.1 | 0.2 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.1 | 1.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 14.0 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 0.7 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.2 | GO:0031652 | positive regulation of heat generation(GO:0031652) |
| 0.1 | 0.4 | GO:0002587 | negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.1 | 3.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.1 | GO:0097114 | NMDA glutamate receptor clustering(GO:0097114) |
| 0.1 | 0.3 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.1 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 0.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 2.5 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 2.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 1.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:1990791 | negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) dorsal root ganglion development(GO:1990791) |
| 0.1 | 0.8 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.3 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 0.3 | GO:0070342 | brown fat cell proliferation(GO:0070342) regulation of brown fat cell proliferation(GO:0070347) |
| 0.1 | 0.2 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.6 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 0.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.2 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 1.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.1 | 0.8 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 1.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.1 | 0.5 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.1 | 1.2 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.4 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.4 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 2.2 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 | 0.2 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.9 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 2.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.5 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) |
| 0.1 | 0.7 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.1 | GO:2000538 | signal complex assembly(GO:0007172) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 1.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.1 | GO:2000790 | regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.1 | 0.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 0.8 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.8 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.3 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.3 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.3 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 1.2 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 0.1 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) positive regulation of glial cell migration(GO:1903977) |
| 0.1 | 0.5 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.2 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.4 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.6 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 1.0 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 3.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 0.3 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.6 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 0.1 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 1.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.6 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 1.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 0.1 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 0.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.2 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.1 | GO:1903421 | regulation of synaptic vesicle recycling(GO:1903421) |
| 0.1 | 1.8 | GO:0032743 | positive regulation of interleukin-2 production(GO:0032743) |
| 0.1 | 1.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.6 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 1.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.2 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.2 | GO:1903624 | regulation of DNA catabolic process(GO:1903624) |
| 0.1 | 0.3 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.1 | 0.3 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.3 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.1 | 0.1 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.1 | 0.2 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 0.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.4 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 1.4 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.1 | 1.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.6 | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.1 | 0.5 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.8 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.8 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.2 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) regulation of MDA-5 signaling pathway(GO:0039533) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 2.4 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.1 | 0.2 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.1 | 0.2 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.5 | GO:0071294 | synaptic transmission, glycinergic(GO:0060012) cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 4.4 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0071223 | response to lipoteichoic acid(GO:0070391) cellular response to lipoteichoic acid(GO:0071223) |
| 0.0 | 0.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 2.0 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 1.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.6 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.2 | GO:0019471 | 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 1.9 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.8 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 5.2 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.2 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.6 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.3 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.0 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.2 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 2.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.5 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0046340 | long-chain fatty acid catabolic process(GO:0042758) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.3 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 3.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:1901142 | insulin metabolic process(GO:1901142) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.4 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 0.2 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.6 | GO:0050856 | regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 1.6 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0001806 | type IV hypersensitivity(GO:0001806) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0033605 | positive regulation of catecholamine secretion(GO:0033605) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 1.1 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.5 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:2000418 | regulation of eosinophil migration(GO:2000416) positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.5 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.1 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.6 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.5 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.1 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0048207 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.3 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.2 | GO:1904752 | vascular associated smooth muscle cell migration(GO:1904738) regulation of vascular associated smooth muscle cell migration(GO:1904752) positive regulation of vascular associated smooth muscle cell migration(GO:1904754) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.0 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.2 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.5 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 1.6 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.1 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.3 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.5 | GO:0042462 | eye photoreceptor cell differentiation(GO:0001754) eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.0 | 0.6 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 1.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.1 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.5 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.0 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.0 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.4 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 2.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.5 | GO:1990267 | response to transition metal nanoparticle(GO:1990267) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.1 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.5 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 1.0 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 0.1 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) |
| 0.0 | 0.2 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:2000778 | positive regulation of interleukin-6 secretion(GO:2000778) |
| 0.0 | 0.0 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0061156 | pulmonary artery morphogenesis(GO:0061156) |
| 0.0 | 0.4 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.0 | 0.1 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 0.0 | GO:1903365 | regulation of fear response(GO:1903365) positive regulation of fear response(GO:1903367) regulation of behavioral fear response(GO:2000822) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.0 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.3 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 11.3 | GO:0030478 | actin cap(GO:0030478) |
| 1.2 | 3.6 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.5 | 12.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.4 | 3.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 4.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 5.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 1.6 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.3 | 6.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 3.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 2.0 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.2 | 1.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 0.9 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 4.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.2 | 2.0 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 1.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 1.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 0.7 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 0.9 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.2 | 0.8 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.2 | 0.8 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 4.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 1.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 3.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 5.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.7 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.8 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.5 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 1.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.2 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 6.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.3 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 0.8 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.8 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 2.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.5 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 3.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 2.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.5 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.6 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 1.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.9 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 1.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 3.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 1.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.8 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.3 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.1 | 0.6 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.1 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 0.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 2.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.5 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.1 | 7.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.2 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.1 | 1.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 3.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 3.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.9 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 1.3 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 14.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 3.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 1.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 3.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 1.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0097361 | MMXD complex(GO:0071817) CIA complex(GO:0097361) |
| 0.0 | 0.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 6.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.0 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 1.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 2.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 22.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.0 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 3.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 2.5 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 17.5 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 3.0 | 17.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.8 | 8.8 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.6 | 6.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.5 | 5.9 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.3 | 3.8 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.2 | 7.3 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 1.1 | 3.4 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.1 | 5.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.1 | 11.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.9 | 2.8 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.8 | 4.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.7 | 3.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.7 | 2.7 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.7 | 3.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.6 | 1.8 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.6 | 5.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.6 | 2.9 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.5 | 1.6 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.5 | 2.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.5 | 2.9 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.5 | 1.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.5 | 4.3 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.5 | 2.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.4 | 2.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.4 | 1.3 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 3.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.4 | 1.2 | GO:0004771 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.4 | 2.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.4 | 10.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.4 | 3.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 1.5 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.4 | 1.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.3 | 2.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.3 | 1.0 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.3 | 1.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 0.9 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.3 | 0.9 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.3 | 1.3 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.3 | 4.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 5.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.3 | 1.5 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.3 | 0.9 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.3 | 1.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 0.8 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.3 | 3.0 | GO:0046977 | TAP binding(GO:0046977) |
| 0.3 | 0.3 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.3 | 0.8 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 1.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 1.0 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.2 | 0.7 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.2 | 1.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 0.6 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.2 | 1.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 1.0 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.2 | 0.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 2.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 0.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 3.9 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 0.9 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 1.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.9 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 1.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 0.7 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.2 | 0.5 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 0.7 | GO:0031711 | tripeptidyl-peptidase activity(GO:0008240) bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.2 | 1.0 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 1.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 0.8 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.5 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 0.5 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 0.8 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 1.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 2.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 2.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 3.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 3.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 1.0 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 1.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.4 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.7 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 1.5 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.1 | 1.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 2.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.8 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.9 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.6 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.9 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 0.8 | GO:0004528 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.7 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 1.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.6 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 1.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.6 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 2.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 2.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 0.5 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.6 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 2.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.3 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 3.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.5 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.4 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 3.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 2.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.3 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 2.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.7 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 2.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 3.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.3 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 3.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 1.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.4 | GO:0052796 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.2 | GO:0099583 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 1.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.8 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.1 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.3 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.1 | 0.4 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 2.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.2 | GO:0047237 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.1 | 14.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.9 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.5 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.9 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 2.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 1.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 1.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 1.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.2 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 0.9 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 0.2 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.1 | 0.7 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 2.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 2.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.5 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 2.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 4.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.5 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 5.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 2.1 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 1.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.3 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.1 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.0 | 0.1 | GO:0099589 | serotonin receptor activity(GO:0099589) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 4.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0015108 | chloride transmembrane transporter activity(GO:0015108) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.9 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 1.6 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.0 | 0.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.3 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 13.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 6.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 5.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 7.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 9.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 5.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 3.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 7.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 5.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 3.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.5 | 1.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.5 | 6.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.5 | 3.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.4 | 8.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 6.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 12.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 4.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 4.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 2.9 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 4.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 3.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 2.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 3.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 4.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 6.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 3.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 2.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.9 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.3 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 2.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 3.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 3.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 10.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 4.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.5 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.6 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.8 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 2.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.2 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 1.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 1.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.8 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 2.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 2.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 4.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.2 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |