Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
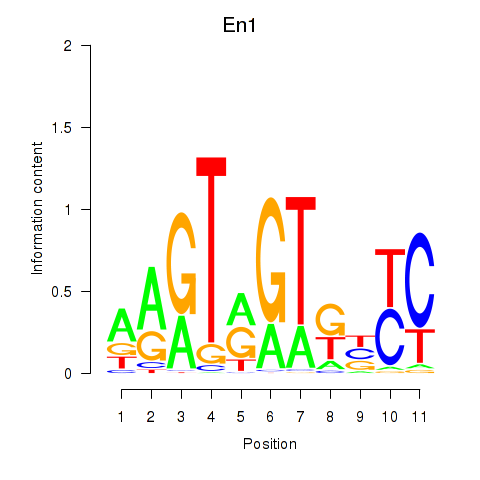
Results for En1
Z-value: 1.01
Transcription factors associated with En1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
En1
|
ENSMUSG00000058665.7 | engrailed 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| En1 | mm10_v2_chr1_+_120602405_120602418 | 0.11 | 5.3e-01 | Click! |
Activity profile of En1 motif
Sorted Z-values of En1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_81975742 | 9.21 |
ENSMUST00000029645.8
|
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr8_+_105048592 | 7.49 |
ENSMUST00000093222.5
ENSMUST00000093223.3 |
Ces3a
|
carboxylesterase 3A |
| chr10_-_128960965 | 7.09 |
ENSMUST00000026398.3
|
Mettl7b
|
methyltransferase like 7B |
| chr1_+_72824482 | 5.59 |
ENSMUST00000047328.4
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr17_-_84682932 | 5.00 |
ENSMUST00000066175.3
|
Abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr4_-_49383576 | 4.94 |
ENSMUST00000107698.1
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr6_-_141856171 | 4.90 |
ENSMUST00000165990.1
ENSMUST00000163678.1 |
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr3_+_130617645 | 4.24 |
ENSMUST00000163620.1
|
Etnppl
|
ethanolamine phosphate phospholyase |
| chr8_+_67490758 | 4.06 |
ENSMUST00000026677.3
|
Nat1
|
N-acetyl transferase 1 |
| chr14_-_51146757 | 4.02 |
ENSMUST00000080126.2
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr17_+_84683131 | 3.97 |
ENSMUST00000171915.1
|
Abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr1_+_88211956 | 3.95 |
ENSMUST00000073049.6
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr13_+_4436094 | 3.90 |
ENSMUST00000156277.1
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr3_+_62419668 | 3.88 |
ENSMUST00000161057.1
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr4_-_62150810 | 3.84 |
ENSMUST00000077719.3
|
Mup21
|
major urinary protein 21 |
| chr5_-_87569023 | 3.74 |
ENSMUST00000113314.2
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr19_-_7802578 | 3.57 |
ENSMUST00000120522.1
ENSMUST00000065634.7 |
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr7_-_68275098 | 3.57 |
ENSMUST00000135564.1
|
Gm16157
|
predicted gene 16157 |
| chr18_+_20247340 | 3.55 |
ENSMUST00000054128.6
|
Dsg1c
|
desmoglein 1 gamma |
| chr10_+_128254131 | 3.45 |
ENSMUST00000060782.3
|
Apon
|
apolipoprotein N |
| chr13_+_4059565 | 3.44 |
ENSMUST00000041768.6
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr3_-_75270073 | 3.31 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr15_+_4727202 | 3.13 |
ENSMUST00000161997.1
ENSMUST00000022788.8 |
C6
|
complement component 6 |
| chr15_+_4727175 | 3.09 |
ENSMUST00000162585.1
|
C6
|
complement component 6 |
| chr1_+_67123015 | 3.03 |
ENSMUST00000027144.7
|
Cps1
|
carbamoyl-phosphate synthetase 1 |
| chr10_+_62071014 | 3.00 |
ENSMUST00000053865.5
|
Gm5424
|
predicted gene 5424 |
| chr16_-_46010212 | 2.86 |
ENSMUST00000130481.1
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr15_+_4727265 | 2.82 |
ENSMUST00000162350.1
|
C6
|
complement component 6 |
| chr13_-_71963713 | 2.71 |
ENSMUST00000077337.8
|
Irx1
|
Iroquois related homeobox 1 (Drosophila) |
| chr5_-_87254804 | 2.70 |
ENSMUST00000075858.3
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr7_+_68275970 | 2.60 |
ENSMUST00000153805.1
|
Fam169b
|
family with sequence similarity 169, member B |
| chr1_-_13660476 | 2.60 |
ENSMUST00000027071.5
|
Lactb2
|
lactamase, beta 2 |
| chr15_+_10177623 | 2.57 |
ENSMUST00000124470.1
|
Prlr
|
prolactin receptor |
| chr4_+_43631935 | 2.55 |
ENSMUST00000030191.8
|
Npr2
|
natriuretic peptide receptor 2 |
| chr12_+_104406704 | 2.54 |
ENSMUST00000021506.5
|
Serpina3n
|
serine (or cysteine) peptidase inhibitor, clade A, member 3N |
| chr17_+_12378603 | 2.52 |
ENSMUST00000014578.5
|
Plg
|
plasminogen |
| chr3_-_113532288 | 2.49 |
ENSMUST00000132353.1
|
Amy2a1
|
amylase 2a1 |
| chr1_-_162898665 | 2.47 |
ENSMUST00000111510.1
ENSMUST00000045902.6 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr8_+_123212857 | 2.42 |
ENSMUST00000060133.6
|
Spata33
|
spermatogenesis associated 33 |
| chr3_-_86999284 | 2.41 |
ENSMUST00000063869.5
ENSMUST00000029717.2 |
Cd1d1
|
CD1d1 antigen |
| chr12_+_8012359 | 2.41 |
ENSMUST00000171239.1
|
Apob
|
apolipoprotein B |
| chr4_-_59960659 | 2.32 |
ENSMUST00000075973.2
|
Mup4
|
major urinary protein 4 |
| chr14_+_52824340 | 2.31 |
ENSMUST00000103648.2
|
Trav11d
|
T cell receptor alpha variable 11D |
| chr9_+_57697612 | 2.29 |
ENSMUST00000034865.4
|
Cyp1a1
|
cytochrome P450, family 1, subfamily a, polypeptide 1 |
| chr4_+_43632185 | 2.27 |
ENSMUST00000107874.2
|
Npr2
|
natriuretic peptide receptor 2 |
| chr1_+_88138364 | 2.18 |
ENSMUST00000014263.4
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr19_+_41029275 | 2.17 |
ENSMUST00000051806.4
ENSMUST00000112200.1 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr12_-_98577940 | 2.16 |
ENSMUST00000110113.1
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chr7_-_26939377 | 2.16 |
ENSMUST00000170227.1
|
Cyp2a22
|
cytochrome P450, family 2, subfamily a, polypeptide 22 |
| chr14_-_66124482 | 2.14 |
ENSMUST00000070515.1
|
Ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr3_+_94693556 | 2.09 |
ENSMUST00000090848.3
ENSMUST00000173981.1 ENSMUST00000173849.1 ENSMUST00000174223.1 |
Selenbp2
|
selenium binding protein 2 |
| chr7_-_27333602 | 2.05 |
ENSMUST00000118583.1
ENSMUST00000118961.1 ENSMUST00000121175.1 |
Ltbp4
|
latent transforming growth factor beta binding protein 4 |
| chrX_-_75875101 | 2.01 |
ENSMUST00000114059.3
|
Pls3
|
plastin 3 (T-isoform) |
| chr5_-_87092546 | 1.99 |
ENSMUST00000132667.1
ENSMUST00000145617.1 ENSMUST00000094649.4 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr5_-_86518578 | 1.93 |
ENSMUST00000134179.1
|
Tmprss11g
|
transmembrane protease, serine 11g |
| chr1_+_88134786 | 1.89 |
ENSMUST00000113134.1
ENSMUST00000140092.1 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr11_+_97685794 | 1.88 |
ENSMUST00000107584.1
ENSMUST00000107585.2 |
Cisd3
|
CDGSH iron sulfur domain 3 |
| chr17_-_34287770 | 1.88 |
ENSMUST00000174751.1
ENSMUST00000040655.6 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr1_+_87594545 | 1.83 |
ENSMUST00000165109.1
ENSMUST00000070898.5 |
Neu2
|
neuraminidase 2 |
| chr11_+_105975204 | 1.81 |
ENSMUST00000001964.7
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr11_+_82115180 | 1.81 |
ENSMUST00000009329.2
|
Ccl8
|
chemokine (C-C motif) ligand 8 |
| chr1_+_88095054 | 1.80 |
ENSMUST00000150634.1
ENSMUST00000058237.7 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr4_-_76344227 | 1.79 |
ENSMUST00000050757.9
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr18_-_15403680 | 1.76 |
ENSMUST00000079081.6
|
Aqp4
|
aquaporin 4 |
| chrX_-_75874536 | 1.75 |
ENSMUST00000033547.7
|
Pls3
|
plastin 3 (T-isoform) |
| chr1_+_172698046 | 1.72 |
ENSMUST00000038495.3
|
Crp
|
C-reactive protein, pentraxin-related |
| chr4_-_129227883 | 1.68 |
ENSMUST00000106051.1
|
C77080
|
expressed sequence C77080 |
| chr7_+_30169861 | 1.68 |
ENSMUST00000085668.4
|
Gm5113
|
predicted gene 5113 |
| chr13_-_56548534 | 1.67 |
ENSMUST00000062806.4
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr7_-_30973367 | 1.65 |
ENSMUST00000108116.3
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr7_+_65693417 | 1.65 |
ENSMUST00000032726.7
ENSMUST00000107495.3 |
Tm2d3
|
TM2 domain containing 3 |
| chr11_+_28853189 | 1.64 |
ENSMUST00000020759.5
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1 |
| chr11_-_101171302 | 1.64 |
ENSMUST00000164474.1
ENSMUST00000043397.7 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr4_-_41741301 | 1.63 |
ENSMUST00000071561.6
ENSMUST00000059354.8 |
Sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr2_-_52335134 | 1.60 |
ENSMUST00000075301.3
|
Neb
|
nebulin |
| chr19_-_32061438 | 1.59 |
ENSMUST00000096119.4
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr7_-_127935429 | 1.59 |
ENSMUST00000141385.1
ENSMUST00000156152.1 |
Prss36
|
protease, serine, 36 |
| chr7_-_141010759 | 1.57 |
ENSMUST00000026565.6
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr1_+_191575721 | 1.57 |
ENSMUST00000045450.5
|
Ints7
|
integrator complex subunit 7 |
| chr18_+_50128200 | 1.56 |
ENSMUST00000025385.6
|
Hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr2_+_30266721 | 1.55 |
ENSMUST00000113645.1
ENSMUST00000133877.1 ENSMUST00000139719.1 ENSMUST00000113643.1 ENSMUST00000150695.1 |
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr19_-_6942406 | 1.55 |
ENSMUST00000099782.3
|
Gpr137
|
G protein-coupled receptor 137 |
| chr2_+_30266513 | 1.53 |
ENSMUST00000091132.6
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr14_+_33941021 | 1.52 |
ENSMUST00000100720.1
|
Gdf2
|
growth differentiation factor 2 |
| chr7_-_3249711 | 1.49 |
ENSMUST00000108653.2
|
Nlrp12
|
NLR family, pyrin domain containing 12 |
| chr6_-_83677807 | 1.47 |
ENSMUST00000037882.6
|
Cd207
|
CD207 antigen |
| chr17_-_34862473 | 1.46 |
ENSMUST00000025229.4
ENSMUST00000176203.2 ENSMUST00000128767.1 |
Cfb
|
complement factor B |
| chr9_+_72958785 | 1.45 |
ENSMUST00000098567.2
ENSMUST00000034734.8 |
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 homolog (human) |
| chr13_+_19342154 | 1.45 |
ENSMUST00000103566.3
|
Tcrg-C4
|
T cell receptor gamma, constant 4 |
| chr6_+_113471481 | 1.45 |
ENSMUST00000113062.1
|
Il17rc
|
interleukin 17 receptor C |
| chr11_-_72795801 | 1.44 |
ENSMUST00000079681.5
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr5_+_23787691 | 1.38 |
ENSMUST00000030852.6
ENSMUST00000120869.1 ENSMUST00000117783.1 ENSMUST00000115113.2 |
Rint1
|
RAD50 interactor 1 |
| chr10_-_92375367 | 1.38 |
ENSMUST00000182870.1
|
Gm20757
|
predicted gene, 20757 |
| chr11_-_72796028 | 1.37 |
ENSMUST00000156294.1
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr1_-_184883218 | 1.37 |
ENSMUST00000048308.5
|
C130074G19Rik
|
RIKEN cDNA C130074G19 gene |
| chr8_+_46492789 | 1.34 |
ENSMUST00000110371.1
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr11_+_101552135 | 1.34 |
ENSMUST00000103099.1
|
Nbr1
|
neighbor of Brca1 gene 1 |
| chr9_-_106891870 | 1.34 |
ENSMUST00000160503.1
ENSMUST00000159620.2 ENSMUST00000160978.1 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr6_+_113471427 | 1.34 |
ENSMUST00000058300.7
|
Il17rc
|
interleukin 17 receptor C |
| chr19_-_11266122 | 1.33 |
ENSMUST00000169159.1
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr11_+_108395288 | 1.33 |
ENSMUST00000000049.5
|
Apoh
|
apolipoprotein H |
| chr4_-_148160031 | 1.32 |
ENSMUST00000057907.3
|
Fbxo44
|
F-box protein 44 |
| chr13_+_45078692 | 1.32 |
ENSMUST00000054395.6
|
Gm9817
|
predicted gene 9817 |
| chr14_+_52863299 | 1.31 |
ENSMUST00000103616.3
|
Trav15d-1-dv6d-1
|
T cell receptor alpha variable 15D-1-DV6D-1 |
| chr9_-_56928350 | 1.31 |
ENSMUST00000050916.5
|
Snx33
|
sorting nexin 33 |
| chr6_+_124493101 | 1.30 |
ENSMUST00000049124.9
|
C1rl
|
complement component 1, r subcomponent-like |
| chr7_-_30598863 | 1.30 |
ENSMUST00000108150.1
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr11_-_120453473 | 1.29 |
ENSMUST00000026452.2
|
Pde6g
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr8_+_46010596 | 1.29 |
ENSMUST00000110381.2
|
Lrp2bp
|
Lrp2 binding protein |
| chr11_+_70092705 | 1.29 |
ENSMUST00000124721.1
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr17_-_47421873 | 1.28 |
ENSMUST00000073143.6
|
1700001C19Rik
|
RIKEN cDNA 1700001C19 gene |
| chrX_-_162643575 | 1.28 |
ENSMUST00000101102.1
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr11_+_70092653 | 1.27 |
ENSMUST00000143772.1
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr9_+_45182034 | 1.26 |
ENSMUST00000164650.1
|
BC049352
|
cDNA sequence BC049352 |
| chr17_+_69969217 | 1.26 |
ENSMUST00000060072.5
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr3_+_108653931 | 1.24 |
ENSMUST00000029483.8
ENSMUST00000124384.1 |
Clcc1
|
chloride channel CLIC-like 1 |
| chr9_+_122888471 | 1.24 |
ENSMUST00000063980.6
|
Zkscan7
|
zinc finger with KRAB and SCAN domains 7 |
| chr2_+_25456830 | 1.23 |
ENSMUST00000114265.2
ENSMUST00000102918.2 |
Clic3
|
chloride intracellular channel 3 |
| chrX_-_147429189 | 1.22 |
ENSMUST00000033646.2
|
Il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr7_-_127936097 | 1.22 |
ENSMUST00000150591.1
|
Prss36
|
protease, serine, 36 |
| chr17_-_34862122 | 1.22 |
ENSMUST00000154526.1
|
Cfb
|
complement factor B |
| chrX_-_100412587 | 1.21 |
ENSMUST00000033567.8
|
Awat2
|
acyl-CoA wax alcohol acyltransferase 2 |
| chr3_+_108653907 | 1.19 |
ENSMUST00000106609.1
|
Clcc1
|
chloride channel CLIC-like 1 |
| chr16_-_45953493 | 1.19 |
ENSMUST00000136405.1
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr7_-_31110997 | 1.17 |
ENSMUST00000039435.8
|
Hpn
|
hepsin |
| chr9_-_40006947 | 1.17 |
ENSMUST00000073932.3
|
Olfr980
|
olfactory receptor 980 |
| chr4_-_148159571 | 1.16 |
ENSMUST00000167160.1
ENSMUST00000151246.1 |
Fbxo44
|
F-box protein 44 |
| chr4_-_42168603 | 1.16 |
ENSMUST00000098121.3
|
Gm13305
|
predicted gene 13305 |
| chr9_-_59750616 | 1.15 |
ENSMUST00000163586.1
ENSMUST00000177963.1 ENSMUST00000051039.4 |
Senp8
|
SUMO/sentrin specific peptidase 8 |
| chr16_+_20548577 | 1.15 |
ENSMUST00000003319.5
|
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr1_+_183418503 | 1.15 |
ENSMUST00000065900.4
|
Hhipl2
|
hedgehog interacting protein-like 2 |
| chr13_+_67833235 | 1.14 |
ENSMUST00000060609.7
|
Gm10037
|
predicted gene 10037 |
| chr9_-_64341288 | 1.14 |
ENSMUST00000068367.7
|
Dis3l
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr3_+_96635817 | 1.13 |
ENSMUST00000139739.1
|
Pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr6_-_90224438 | 1.13 |
ENSMUST00000076086.2
|
Vmn1r53
|
vomeronasal 1 receptor 53 |
| chr7_-_31111148 | 1.13 |
ENSMUST00000164929.1
|
Hpn
|
hepsin |
| chr2_-_30194112 | 1.13 |
ENSMUST00000113659.1
ENSMUST00000113660.1 |
Ccbl1
|
cysteine conjugate-beta lyase 1 |
| chr3_+_19957088 | 1.11 |
ENSMUST00000108328.1
|
Cp
|
ceruloplasmin |
| chr7_-_132154717 | 1.09 |
ENSMUST00000033149.4
|
Cpxm2
|
carboxypeptidase X 2 (M14 family) |
| chr11_+_49280150 | 1.09 |
ENSMUST00000078932.1
|
Olfr1393
|
olfactory receptor 1393 |
| chr11_-_101170327 | 1.09 |
ENSMUST00000123864.1
|
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr13_+_24943144 | 1.08 |
ENSMUST00000021773.5
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr14_+_55559993 | 1.07 |
ENSMUST00000117236.1
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr16_-_20730544 | 1.07 |
ENSMUST00000076422.5
|
Thpo
|
thrombopoietin |
| chr1_+_74284930 | 1.07 |
ENSMUST00000113805.1
ENSMUST00000027370.6 ENSMUST00000087226.4 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr6_-_69631933 | 1.07 |
ENSMUST00000177697.1
|
Igkv4-54
|
immunoglobulin kappa chain variable 4-54 |
| chr3_+_96635840 | 1.06 |
ENSMUST00000165842.1
|
Pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr6_-_142278836 | 1.05 |
ENSMUST00000111825.3
|
Slco1a5
|
solute carrier organic anion transporter family, member 1a5 |
| chr3_+_19957037 | 1.04 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr19_+_37685581 | 1.03 |
ENSMUST00000073391.4
|
Cyp26c1
|
cytochrome P450, family 26, subfamily c, polypeptide 1 |
| chr4_+_22357543 | 1.02 |
ENSMUST00000039234.3
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr5_-_88527841 | 1.02 |
ENSMUST00000087033.3
|
Igj
|
immunoglobulin joining chain |
| chr2_+_32606946 | 1.01 |
ENSMUST00000113290.1
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr11_+_69991633 | 1.01 |
ENSMUST00000108592.1
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr16_-_96082389 | 1.01 |
ENSMUST00000099502.2
ENSMUST00000023631.8 ENSMUST00000113829.1 ENSMUST00000153398.1 |
Brwd1
|
bromodomain and WD repeat domain containing 1 |
| chr6_-_3968357 | 1.00 |
ENSMUST00000031674.8
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr16_-_38522662 | 1.00 |
ENSMUST00000002925.5
|
Timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr11_+_70459940 | 1.00 |
ENSMUST00000147289.1
ENSMUST00000126105.1 |
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr4_-_148159838 | 1.00 |
ENSMUST00000151127.1
ENSMUST00000105705.2 |
Fbxo44
|
F-box protein 44 |
| chrM_+_10167 | 1.00 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr7_-_71351485 | 1.00 |
ENSMUST00000094315.2
|
Gm10295
|
predicted gene 10295 |
| chr6_-_28133325 | 1.00 |
ENSMUST00000131897.1
|
Grm8
|
glutamate receptor, metabotropic 8 |
| chrM_+_9452 | 0.99 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr14_-_16243309 | 0.99 |
ENSMUST00000112625.1
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr11_-_69369377 | 0.98 |
ENSMUST00000092971.6
ENSMUST00000108661.1 |
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr2_+_32606979 | 0.97 |
ENSMUST00000113289.1
ENSMUST00000095044.3 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr12_-_119238794 | 0.96 |
ENSMUST00000026360.8
|
Itgb8
|
integrin beta 8 |
| chr5_+_35583018 | 0.96 |
ENSMUST00000068947.7
ENSMUST00000114237.1 ENSMUST00000156125.1 ENSMUST00000068563.5 |
Acox3
|
acyl-Coenzyme A oxidase 3, pristanoyl |
| chr5_-_72752763 | 0.95 |
ENSMUST00000113604.3
|
Txk
|
TXK tyrosine kinase |
| chr3_+_93442330 | 0.95 |
ENSMUST00000064257.5
|
Tchh
|
trichohyalin |
| chr16_-_91618986 | 0.94 |
ENSMUST00000143058.1
ENSMUST00000049244.8 ENSMUST00000169982.1 ENSMUST00000133731.1 |
Dnajc28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr4_+_102421518 | 0.94 |
ENSMUST00000106904.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr2_-_26140468 | 0.94 |
ENSMUST00000133808.1
|
C330006A16Rik
|
RIKEN cDNA C330006A16 gene |
| chr5_-_86676346 | 0.94 |
ENSMUST00000038448.6
|
Tmprss11bnl
|
transmembrane protease, serine 11b N terminal like |
| chr18_+_20376723 | 0.91 |
ENSMUST00000076737.6
|
Dsg1b
|
desmoglein 1 beta |
| chr2_+_14229390 | 0.90 |
ENSMUST00000028045.2
|
Mrc1
|
mannose receptor, C type 1 |
| chr3_+_19957240 | 0.90 |
ENSMUST00000108325.2
|
Cp
|
ceruloplasmin |
| chr13_+_40859768 | 0.89 |
ENSMUST00000110191.2
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr14_+_52753367 | 0.89 |
ENSMUST00000180717.1
ENSMUST00000183820.1 |
Trav6d-4
|
T cell receptor alpha variable 6D-4 |
| chr3_+_114904062 | 0.88 |
ENSMUST00000081752.6
|
Olfm3
|
olfactomedin 3 |
| chr6_+_142298419 | 0.88 |
ENSMUST00000041993.2
|
Iapp
|
islet amyloid polypeptide |
| chr16_-_45953565 | 0.88 |
ENSMUST00000134802.1
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr3_+_123267445 | 0.87 |
ENSMUST00000047923.7
|
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr9_-_64341145 | 0.86 |
ENSMUST00000120760.1
ENSMUST00000168844.2 |
Dis3l
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr17_-_33760306 | 0.86 |
ENSMUST00000173860.1
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr5_-_146220901 | 0.85 |
ENSMUST00000169407.2
ENSMUST00000161331.1 ENSMUST00000159074.2 ENSMUST00000067837.3 |
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr11_+_87595646 | 0.85 |
ENSMUST00000134216.1
|
Mtmr4
|
myotubularin related protein 4 |
| chrX_+_139563316 | 0.85 |
ENSMUST00000113027.1
|
Rnf128
|
ring finger protein 128 |
| chr8_+_22624019 | 0.85 |
ENSMUST00000033936.6
|
Dkk4
|
dickkopf homolog 4 (Xenopus laevis) |
| chr2_+_101624696 | 0.84 |
ENSMUST00000044031.3
|
Rag2
|
recombination activating gene 2 |
| chr16_+_42907563 | 0.84 |
ENSMUST00000151244.1
ENSMUST00000114694.2 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr14_-_30915387 | 0.84 |
ENSMUST00000166622.1
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr11_+_105967938 | 0.83 |
ENSMUST00000001963.7
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr10_-_13553111 | 0.83 |
ENSMUST00000105539.1
|
Pex3
|
peroxisomal biogenesis factor 3 |
| chr7_+_65693447 | 0.83 |
ENSMUST00000143508.1
|
Tm2d3
|
TM2 domain containing 3 |
| chr3_-_117868821 | 0.83 |
ENSMUST00000167877.1
ENSMUST00000169812.1 |
Snx7
|
sorting nexin 7 |
| chr11_-_99374895 | 0.83 |
ENSMUST00000006963.2
|
Krt28
|
keratin 28 |
| chr4_+_88841816 | 0.82 |
ENSMUST00000094973.3
|
Ifna4
|
interferon alpha 4 |
| chr7_+_101321703 | 0.82 |
ENSMUST00000174291.1
ENSMUST00000167888.2 ENSMUST00000172662.1 ENSMUST00000173270.1 ENSMUST00000174083.1 |
Stard10
|
START domain containing 10 |
| chr2_+_91257323 | 0.81 |
ENSMUST00000111349.2
ENSMUST00000131711.1 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of En1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 3.0 | 9.0 | GO:0045796 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 1.8 | 9.0 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.6 | 6.2 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) |
| 1.3 | 3.9 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.1 | 3.4 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.9 | 2.6 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.8 | 2.4 | GO:0048007 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.7 | 3.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.7 | 2.1 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.7 | 2.7 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.6 | 2.3 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.6 | 1.7 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.5 | 2.5 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.5 | 1.5 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.5 | 4.9 | GO:1900194 | receptor guanylyl cyclase signaling pathway(GO:0007168) negative regulation of oocyte maturation(GO:1900194) |
| 0.5 | 1.4 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.4 | 3.0 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.4 | 2.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.4 | 4.1 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.4 | 2.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.4 | 1.1 | GO:0006507 | GPI anchor release(GO:0006507) |
| 0.4 | 1.8 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.3 | 2.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 1.0 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.3 | 1.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 1.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 1.5 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.3 | 1.7 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.3 | 1.7 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.3 | 0.3 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.3 | 1.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.3 | 1.6 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.3 | 1.0 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.3 | 1.0 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.3 | 1.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.3 | 3.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 0.7 | GO:0030862 | neuroblast division in subventricular zone(GO:0021849) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.2 | 2.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 1.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.2 | 0.7 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) epicardium-derived cardiac vascular smooth muscle cell differentiation(GO:0060983) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.2 | 1.2 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.2 | 1.8 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 0.7 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 1.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 0.4 | GO:1903093 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.2 | 0.6 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.2 | 0.6 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.2 | 2.9 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.2 | 1.2 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 0.8 | GO:1902956 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.2 | 6.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 5.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 2.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.3 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.2 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 2.0 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.2 | 0.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 3.9 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 1.8 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.2 | 1.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 1.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 0.5 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.2 | 0.6 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.2 | 1.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 2.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.8 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 3.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 1.2 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 2.0 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 3.9 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.7 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.1 | 0.9 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.7 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 2.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.6 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.2 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.4 | GO:0036395 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 0.1 | GO:2000229 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.1 | 0.9 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.9 | GO:0097647 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 1.6 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.1 | 0.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 0.3 | GO:1990167 | protein K6-linked deubiquitination(GO:0044313) protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 1.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.4 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.1 | 1.0 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.3 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 1.3 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.8 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 0.7 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.5 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.3 | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) |
| 0.1 | 1.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 2.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.3 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 1.0 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 5.8 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.1 | 0.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.7 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 1.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.9 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 1.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.5 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.1 | 0.7 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.4 | GO:0021917 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.1 | 0.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 2.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.6 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 1.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.3 | GO:1903416 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) response to glycoside(GO:1903416) |
| 0.1 | 1.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 2.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.3 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.1 | 0.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.5 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.1 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.2 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.1 | 2.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 0.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 1.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.3 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 2.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 4.4 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.4 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.1 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.3 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 1.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 1.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.8 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.1 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.4 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 4.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 1.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.7 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.3 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.8 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.0 | 0.7 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.8 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.4 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.9 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 3.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 2.6 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.4 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 3.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.8 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.5 | GO:0071384 | cellular response to corticosteroid stimulus(GO:0071384) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.8 | 2.5 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.8 | 9.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 1.4 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.3 | 3.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 5.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 0.6 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 2.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 2.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 0.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 3.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 4.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 2.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 4.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.8 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.1 | 3.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.7 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.7 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 1.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 3.0 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.6 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.7 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 9.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 3.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.3 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.9 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.9 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 1.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 2.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:1990131 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 3.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 3.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.0 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 4.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 1.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 2.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 1.4 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 3.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 3.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.4 | 4.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 1.1 | 6.9 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 1.0 | 4.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 1.0 | 3.0 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 1.0 | 4.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.7 | 3.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.7 | 2.6 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) metallodipeptidase activity(GO:0070573) |
| 0.6 | 5.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.6 | 4.0 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.5 | 2.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.4 | 2.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.4 | 1.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 1.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.4 | 2.4 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.4 | 1.6 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.4 | 14.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 11.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.4 | 1.8 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 2.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 3.1 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.3 | 1.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 1.3 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.3 | 6.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.3 | 2.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 1.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.3 | 1.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 1.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 1.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 1.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 1.0 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.2 | 2.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 3.6 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 0.9 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 1.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 2.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 1.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.2 | 0.6 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.2 | 2.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.2 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.2 | 0.8 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.6 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.2 | 0.5 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.1 | 0.7 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.4 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 1.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 4.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 2.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.6 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.1 | 2.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 1.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 1.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.7 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 1.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.3 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.8 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 2.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 1.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.5 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 1.0 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 1.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 2.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.4 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 1.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.8 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 1.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 1.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 2.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.3 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 7.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 1.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 1.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.6 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.6 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 1.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 5.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 5.5 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.7 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 1.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 1.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.6 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 2.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 5.4 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 1.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 4.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 3.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 2.3 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 4.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 3.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 4.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 7.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.6 | 10.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.5 | 11.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 13.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 7.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 3.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 2.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 3.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 2.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 0.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 2.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 4.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.9 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |