Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Egr3
Z-value: 0.91
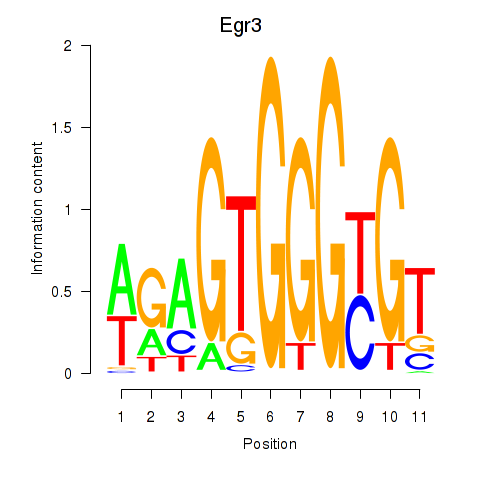
Transcription factors associated with Egr3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Egr3
|
ENSMUSG00000033730.3 | early growth response 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr3 | mm10_v2_chr14_+_70077375_70077445 | -0.10 | 5.7e-01 | Click! |
Activity profile of Egr3 motif
Sorted Z-values of Egr3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_81975742 | 9.73 |
ENSMUST00000029645.8
|
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr13_-_41847626 | 5.52 |
ENSMUST00000121404.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr13_-_41847599 | 4.95 |
ENSMUST00000179758.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr13_-_41847482 | 4.92 |
ENSMUST00000072012.3
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr10_-_68278713 | 4.33 |
ENSMUST00000020106.7
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr2_-_148045891 | 3.50 |
ENSMUST00000109964.1
|
Foxa2
|
forkhead box A2 |
| chr6_-_124542281 | 3.20 |
ENSMUST00000159463.1
ENSMUST00000162844.1 ENSMUST00000160505.1 ENSMUST00000162443.1 |
C1s
|
complement component 1, s subcomponent |
| chr11_-_98775333 | 2.87 |
ENSMUST00000064941.6
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr3_-_138131356 | 2.50 |
ENSMUST00000029805.8
|
Mttp
|
microsomal triglyceride transfer protein |
| chr2_-_173119402 | 2.24 |
ENSMUST00000094287.3
ENSMUST00000179693.1 |
Ctcfl
|
CCCTC-binding factor (zinc finger protein)-like |
| chr6_+_91684061 | 2.23 |
ENSMUST00000032185.7
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr4_-_57300362 | 2.23 |
ENSMUST00000153926.1
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr8_-_124751808 | 2.13 |
ENSMUST00000055257.5
|
Fam89a
|
family with sequence similarity 89, member A |
| chr19_-_38124801 | 2.10 |
ENSMUST00000112335.2
|
Rbp4
|
retinol binding protein 4, plasma |
| chr7_+_80246529 | 2.10 |
ENSMUST00000107381.1
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr7_+_80246375 | 2.05 |
ENSMUST00000058266.6
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr2_+_116067213 | 2.04 |
ENSMUST00000152412.1
|
G630016G05Rik
|
RIKEN cDNA G630016G05 gene |
| chr17_-_87282793 | 1.89 |
ENSMUST00000146560.2
|
4833418N02Rik
|
RIKEN cDNA 4833418N02 gene |
| chr11_-_109722214 | 1.85 |
ENSMUST00000020938.7
|
Fam20a
|
family with sequence similarity 20, member A |
| chr10_+_29143996 | 1.85 |
ENSMUST00000092629.2
|
Soga3
|
SOGA family member 3 |
| chr1_-_157412576 | 1.70 |
ENSMUST00000078308.6
ENSMUST00000139470.1 |
Rasal2
|
RAS protein activator like 2 |
| chr7_+_128523576 | 1.68 |
ENSMUST00000033136.7
|
Bag3
|
BCL2-associated athanogene 3 |
| chr18_-_39490649 | 1.59 |
ENSMUST00000115567.1
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr2_+_32395896 | 1.55 |
ENSMUST00000028162.3
|
Ptges2
|
prostaglandin E synthase 2 |
| chr10_+_59221945 | 1.55 |
ENSMUST00000182161.1
|
Sowahc
|
sosondowah ankyrin repeat domain family member C |
| chr16_+_64851991 | 1.53 |
ENSMUST00000067744.7
|
Cggbp1
|
CGG triplet repeat binding protein 1 |
| chr6_-_144209558 | 1.53 |
ENSMUST00000111749.1
ENSMUST00000170367.2 |
Sox5
|
SRY-box containing gene 5 |
| chr7_-_19023538 | 1.41 |
ENSMUST00000036018.5
|
Foxa3
|
forkhead box A3 |
| chr5_+_102481546 | 1.41 |
ENSMUST00000112854.1
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr4_-_41695935 | 1.40 |
ENSMUST00000145379.1
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr18_-_37935403 | 1.40 |
ENSMUST00000080033.6
ENSMUST00000115631.1 |
Diap1
|
diaphanous homolog 1 (Drosophila) |
| chr18_-_37935378 | 1.40 |
ENSMUST00000025337.7
|
Diap1
|
diaphanous homolog 1 (Drosophila) |
| chr5_-_62766153 | 1.40 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr11_+_98937669 | 1.37 |
ENSMUST00000107475.2
ENSMUST00000068133.3 |
Rara
|
retinoic acid receptor, alpha |
| chr2_+_121413775 | 1.37 |
ENSMUST00000028683.7
|
Pdia3
|
protein disulfide isomerase associated 3 |
| chr18_-_37935429 | 1.36 |
ENSMUST00000115634.1
|
Diap1
|
diaphanous homolog 1 (Drosophila) |
| chr9_-_75409951 | 1.35 |
ENSMUST00000049355.10
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr9_+_44499126 | 1.30 |
ENSMUST00000074989.5
|
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr7_-_119184374 | 1.28 |
ENSMUST00000084650.4
|
Gpr139
|
G protein-coupled receptor 139 |
| chr7_-_141539784 | 1.28 |
ENSMUST00000118694.1
ENSMUST00000153191.1 ENSMUST00000166082.1 ENSMUST00000026586.6 |
Chid1
|
chitinase domain containing 1 |
| chr17_-_87282771 | 1.26 |
ENSMUST00000161759.1
|
4833418N02Rik
|
RIKEN cDNA 4833418N02 gene |
| chr7_+_48959089 | 1.25 |
ENSMUST00000183659.1
|
Nav2
|
neuron navigator 2 |
| chr1_+_120340569 | 1.24 |
ENSMUST00000037286.8
|
C1ql2
|
complement component 1, q subcomponent-like 2 |
| chr9_+_75410145 | 1.17 |
ENSMUST00000180533.1
ENSMUST00000180574.1 |
4933433G15Rik
|
RIKEN cDNA 4933433G15 gene |
| chr3_+_135438722 | 1.16 |
ENSMUST00000166033.1
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr5_-_62765618 | 1.14 |
ENSMUST00000159470.1
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr7_+_3693602 | 1.10 |
ENSMUST00000123088.1
ENSMUST00000038521.7 ENSMUST00000108629.1 ENSMUST00000142713.1 |
Tsen34
|
tRNA splicing endonuclease 34 homolog (S. cerevisiae) |
| chr1_-_9299238 | 1.10 |
ENSMUST00000140295.1
|
Sntg1
|
syntrophin, gamma 1 |
| chr4_+_57637816 | 1.09 |
ENSMUST00000150412.1
|
Gm20459
|
predicted gene 20459 |
| chrX_+_143664365 | 1.08 |
ENSMUST00000126592.1
ENSMUST00000156449.1 ENSMUST00000155215.1 ENSMUST00000112865.1 |
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr6_+_42261957 | 1.07 |
ENSMUST00000095987.3
|
Tmem139
|
transmembrane protein 139 |
| chr17_-_79715034 | 1.07 |
ENSMUST00000024894.1
|
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr7_-_144939823 | 1.06 |
ENSMUST00000093962.4
|
Ccnd1
|
cyclin D1 |
| chr5_+_64970069 | 1.05 |
ENSMUST00000031080.8
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chr8_+_19194319 | 0.98 |
ENSMUST00000052601.3
|
Defb14
|
defensin beta 14 |
| chr18_-_84086379 | 0.97 |
ENSMUST00000060303.8
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr11_+_71750980 | 0.96 |
ENSMUST00000108511.1
|
Wscd1
|
WSC domain containing 1 |
| chr5_-_134747241 | 0.90 |
ENSMUST00000015138.9
|
Eln
|
elastin |
| chrX_+_143664290 | 0.88 |
ENSMUST00000112868.1
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr19_-_47157764 | 0.85 |
ENSMUST00000178630.1
|
Calhm3
|
calcium homeostasis modulator 3 |
| chr4_+_40948401 | 0.82 |
ENSMUST00000030128.5
|
Chmp5
|
charged multivesicular body protein 5 |
| chr6_-_83572429 | 0.81 |
ENSMUST00000068054.7
|
Stambp
|
STAM binding protein |
| chr10_-_59221757 | 0.79 |
ENSMUST00000165971.1
|
Sept10
|
septin 10 |
| chr11_+_71750680 | 0.79 |
ENSMUST00000021168.7
|
Wscd1
|
WSC domain containing 1 |
| chr13_+_59585259 | 0.77 |
ENSMUST00000168367.1
ENSMUST00000022038.7 ENSMUST00000166923.1 |
Naa35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr12_-_67221221 | 0.75 |
ENSMUST00000178814.1
ENSMUST00000179345.2 |
Mdga2
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 MAM domain-containing glycosylphosphatidylinositol anchor protein 2 |
| chr10_-_77799133 | 0.73 |
ENSMUST00000178581.1
|
Gm19668
|
predicted gene, 19668 |
| chr10_-_18023229 | 0.71 |
ENSMUST00000020002.7
|
Abracl
|
ABRA C-terminal like |
| chr18_+_65698253 | 0.70 |
ENSMUST00000115097.1
ENSMUST00000117694.1 |
Oacyl
|
O-acyltransferase like |
| chr18_+_32163073 | 0.69 |
ENSMUST00000096575.3
|
Map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr10_+_38965515 | 0.68 |
ENSMUST00000019992.5
|
Lama4
|
laminin, alpha 4 |
| chr12_+_49382791 | 0.67 |
ENSMUST00000179669.1
|
Foxg1
|
forkhead box G1 |
| chrX_-_162159717 | 0.65 |
ENSMUST00000087085.3
|
Nhs
|
Nance-Horan syndrome (human) |
| chrX_-_48513518 | 0.63 |
ENSMUST00000114945.2
ENSMUST00000037349.7 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr11_-_33276334 | 0.62 |
ENSMUST00000183831.1
|
Gm12117
|
predicted gene 12117 |
| chr7_-_110061319 | 0.62 |
ENSMUST00000098110.2
|
AA474408
|
expressed sequence AA474408 |
| chr19_-_38224096 | 0.60 |
ENSMUST00000067167.5
|
Fra10ac1
|
FRA10AC1 homolog (human) |
| chr4_+_99955715 | 0.60 |
ENSMUST00000102783.4
|
Pgm2
|
phosphoglucomutase 2 |
| chr9_-_78481724 | 0.59 |
ENSMUST00000042235.8
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr7_-_45510400 | 0.57 |
ENSMUST00000033096.7
|
Nucb1
|
nucleobindin 1 |
| chr10_-_128645784 | 0.56 |
ENSMUST00000065334.3
|
Ikzf4
|
IKAROS family zinc finger 4 |
| chr17_-_46327949 | 0.55 |
ENSMUST00000047970.7
|
Abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr10_+_40883819 | 0.55 |
ENSMUST00000105509.1
|
Wasf1
|
WAS protein family, member 1 |
| chr6_+_103510874 | 0.54 |
ENSMUST00000066905.6
|
Chl1
|
cell adhesion molecule with homology to L1CAM |
| chr1_+_74236479 | 0.53 |
ENSMUST00000113820.2
ENSMUST00000006467.7 ENSMUST00000113819.1 |
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr2_-_63184253 | 0.53 |
ENSMUST00000075052.3
ENSMUST00000112454.1 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr17_-_46327990 | 0.53 |
ENSMUST00000167360.1
|
Abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr1_+_86526688 | 0.53 |
ENSMUST00000045897.8
|
Ptma
|
prothymosin alpha |
| chr17_-_33760306 | 0.51 |
ENSMUST00000173860.1
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr5_+_143403819 | 0.50 |
ENSMUST00000110731.2
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr4_-_40948196 | 0.48 |
ENSMUST00000030125.4
ENSMUST00000108089.1 |
Bag1
|
BCL2-associated athanogene 1 |
| chr3_+_97901190 | 0.48 |
ENSMUST00000029476.2
ENSMUST00000122288.1 |
Sec22b
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) |
| chr11_-_23895208 | 0.47 |
ENSMUST00000102863.2
ENSMUST00000020513.3 |
Papolg
|
poly(A) polymerase gamma |
| chr7_+_3645267 | 0.47 |
ENSMUST00000038913.9
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr10_-_40883073 | 0.46 |
ENSMUST00000044166.7
|
Cdc40
|
cell division cycle 40 |
| chr10_-_5922385 | 0.45 |
ENSMUST00000131996.1
ENSMUST00000064225.7 |
Rgs17
|
regulator of G-protein signaling 17 |
| chr13_-_85288999 | 0.44 |
ENSMUST00000109552.2
|
Rasa1
|
RAS p21 protein activator 1 |
| chr14_-_21740395 | 0.43 |
ENSMUST00000120984.2
|
Dusp13
|
dual specificity phosphatase 13 |
| chr10_+_40883469 | 0.40 |
ENSMUST00000019975.7
|
Wasf1
|
WAS protein family, member 1 |
| chr11_-_99422252 | 0.40 |
ENSMUST00000017741.3
|
Krt12
|
keratin 12 |
| chr3_+_93393696 | 0.39 |
ENSMUST00000045912.2
|
Rptn
|
repetin |
| chr5_+_129715488 | 0.37 |
ENSMUST00000119576.1
ENSMUST00000042191.5 ENSMUST00000118420.1 ENSMUST00000154358.1 ENSMUST00000121339.1 ENSMUST00000119604.1 ENSMUST00000136108.1 ENSMUST00000121813.1 ENSMUST00000119985.1 ENSMUST00000138812.1 |
Mrps17
|
mitochondrial ribosomal protein S17 |
| chr17_+_87282880 | 0.33 |
ENSMUST00000041110.5
ENSMUST00000125875.1 |
Ttc7
|
tetratricopeptide repeat domain 7 |
| chr8_+_62951361 | 0.33 |
ENSMUST00000119068.1
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chrX_+_36195968 | 0.32 |
ENSMUST00000115256.1
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chr11_-_77489666 | 0.31 |
ENSMUST00000037593.7
ENSMUST00000092892.3 |
Ankrd13b
|
ankyrin repeat domain 13b |
| chr14_+_15437623 | 0.30 |
ENSMUST00000181388.1
|
B230110C06Rik
|
RIKEN cDNA B230110C06 gene |
| chr16_-_94370994 | 0.30 |
ENSMUST00000113914.1
ENSMUST00000113905.1 |
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr7_-_100371889 | 0.29 |
ENSMUST00000032963.8
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr1_-_164307443 | 0.21 |
ENSMUST00000027866.4
ENSMUST00000120447.1 ENSMUST00000086032.3 |
Blzf1
|
basic leucine zipper nuclear factor 1 |
| chr1_-_161788489 | 0.21 |
ENSMUST00000000834.2
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr17_-_29078953 | 0.21 |
ENSMUST00000133221.1
|
Trp53cor1
|
tumor protein p53 pathway corepressor 1 |
| chr10_-_111010001 | 0.19 |
ENSMUST00000099285.3
ENSMUST00000041723.7 |
Zdhhc17
|
zinc finger, DHHC domain containing 17 |
| chr10_+_69534208 | 0.18 |
ENSMUST00000182439.1
ENSMUST00000092434.5 ENSMUST00000092432.5 ENSMUST00000092431.5 ENSMUST00000054167.8 ENSMUST00000047061.6 |
Ank3
|
ankyrin 3, epithelial |
| chr15_+_43477213 | 0.18 |
ENSMUST00000022962.6
|
Emc2
|
ER membrane protein complex subunit 2 |
| chr7_+_80860909 | 0.14 |
ENSMUST00000132163.1
ENSMUST00000147125.1 |
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr12_+_49383007 | 0.14 |
ENSMUST00000021333.3
|
Foxg1
|
forkhead box G1 |
| chr8_+_104170513 | 0.13 |
ENSMUST00000171018.1
ENSMUST00000167633.1 ENSMUST00000093245.5 ENSMUST00000164076.1 |
Bean1
|
brain expressed, associated with Nedd4, 1 |
| chr10_+_69533803 | 0.11 |
ENSMUST00000182155.1
ENSMUST00000183169.1 ENSMUST00000183148.1 |
Ank3
|
ankyrin 3, epithelial |
| chr10_-_5922341 | 0.11 |
ENSMUST00000117676.1
ENSMUST00000019909.7 |
Rgs17
|
regulator of G-protein signaling 17 |
| chr4_-_129696579 | 0.11 |
ENSMUST00000137640.1
|
Tmem39b
|
transmembrane protein 39b |
| chr4_-_129662442 | 0.08 |
ENSMUST00000003828.4
|
Kpna6
|
karyopherin (importin) alpha 6 |
| chr17_-_83631892 | 0.05 |
ENSMUST00000051482.1
|
Kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr10_+_69533761 | 0.04 |
ENSMUST00000182884.1
|
Ank3
|
ankyrin 3, epithelial |
| chr4_-_126968124 | 0.03 |
ENSMUST00000106108.2
|
Zmym4
|
zinc finger, MYM-type 4 |
| chr7_+_126759601 | 0.02 |
ENSMUST00000050201.4
ENSMUST00000057669.9 |
Mapk3
|
mitogen-activated protein kinase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Egr3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 1.2 | 3.5 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.6 | 2.9 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.5 | 2.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.5 | 4.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.5 | 1.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.5 | 1.4 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.4 | 2.5 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.4 | 1.1 | GO:0071603 | dibenzo-p-dioxin metabolic process(GO:0018894) endothelial cell-cell adhesion(GO:0071603) |
| 0.4 | 1.4 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.4 | 1.4 | GO:0003360 | brainstem development(GO:0003360) |
| 0.3 | 1.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 | 1.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.3 | 0.8 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.3 | 0.8 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.3 | 1.6 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.3 | 1.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 2.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 1.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.5 | GO:2000812 | response to rapamycin(GO:1901355) regulation of barbed-end actin filament capping(GO:2000812) |
| 0.2 | 1.5 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 | 1.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 1.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 1.9 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.9 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.5 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 1.7 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 2.2 | GO:0043046 | regulation of gene expression by genetic imprinting(GO:0006349) DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.6 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 2.0 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 0.3 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.6 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.1 | 1.6 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 2.2 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 2.9 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.4 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.8 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 1.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 1.3 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.6 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 1.4 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.5 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 1.3 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.3 | 0.8 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 0.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 1.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 1.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 2.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.6 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 2.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.5 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.7 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.7 | 2.2 | GO:0030977 | taurine binding(GO:0030977) |
| 0.5 | 1.4 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.4 | 1.6 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.3 | 1.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.3 | 1.6 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 2.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 1.4 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.2 | 1.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 2.1 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.1 | 0.6 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 2.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.4 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 2.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 2.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 1.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 3.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 2.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 1.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 4.1 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 5.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 6.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.6 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 3.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 4.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 3.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 3.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |