Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for E2f7
Z-value: 1.40
Transcription factors associated with E2f7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f7
|
ENSMUSG00000020185.10 | E2F transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f7 | mm10_v2_chr10_+_110745433_110745572 | 0.83 | 5.3e-10 | Click! |
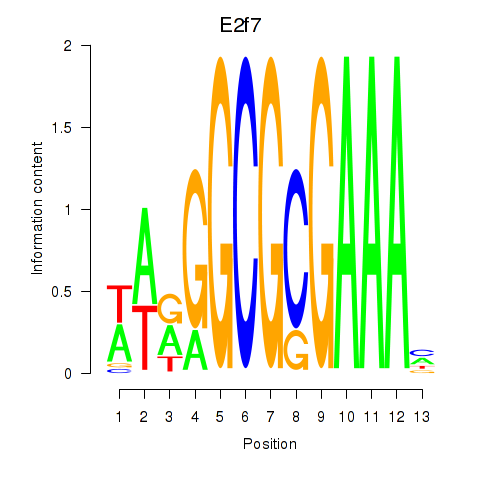
Activity profile of E2f7 motif
Sorted Z-values of E2f7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_108579445 | 15.06 |
ENSMUST00000102744.3
|
Orc1
|
origin recognition complex, subunit 1 |
| chr7_-_142578139 | 11.19 |
ENSMUST00000136359.1
|
H19
|
H19 fetal liver mRNA |
| chr15_+_102296256 | 10.13 |
ENSMUST00000064924.4
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae) |
| chr13_-_100775844 | 9.68 |
ENSMUST00000075550.3
|
Cenph
|
centromere protein H |
| chr12_+_24708984 | 8.71 |
ENSMUST00000154588.1
|
Rrm2
|
ribonucleotide reductase M2 |
| chr10_+_3973086 | 8.65 |
ENSMUST00000117291.1
ENSMUST00000120585.1 ENSMUST00000043735.7 |
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr13_-_21833575 | 8.13 |
ENSMUST00000081342.5
|
Hist1h2ap
|
histone cluster 1, H2ap |
| chr11_+_102248842 | 8.07 |
ENSMUST00000100392.4
|
BC030867
|
cDNA sequence BC030867 |
| chr13_-_21753851 | 7.68 |
ENSMUST00000074752.2
|
Hist1h2ak
|
histone cluster 1, H2ak |
| chr12_+_24708241 | 7.50 |
ENSMUST00000020980.5
|
Rrm2
|
ribonucleotide reductase M2 |
| chr4_+_136172367 | 7.39 |
ENSMUST00000061721.5
|
E2f2
|
E2F transcription factor 2 |
| chr10_-_69352886 | 7.12 |
ENSMUST00000119827.1
ENSMUST00000020099.5 |
Cdk1
|
cyclin-dependent kinase 1 |
| chr3_+_108383829 | 6.10 |
ENSMUST00000090561.3
ENSMUST00000102629.1 ENSMUST00000128089.1 |
Psrc1
|
proline/serine-rich coiled-coil 1 |
| chr9_-_58741543 | 6.08 |
ENSMUST00000098674.4
|
2410076I21Rik
|
RIKEN cDNA 2410076I21 gene |
| chr14_-_54641347 | 5.79 |
ENSMUST00000067784.6
|
Cdh24
|
cadherin-like 24 |
| chr4_+_126556994 | 5.75 |
ENSMUST00000147675.1
|
Clspn
|
claspin |
| chr8_+_75109528 | 5.71 |
ENSMUST00000164309.1
|
Mcm5
|
minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae) |
| chr4_+_126556935 | 5.67 |
ENSMUST00000048391.8
|
Clspn
|
claspin |
| chr2_+_72476159 | 5.39 |
ENSMUST00000102691.4
|
Cdca7
|
cell division cycle associated 7 |
| chr6_-_88898664 | 5.11 |
ENSMUST00000058011.6
|
Mcm2
|
minichromosome maintenance deficient 2 mitotin (S. cerevisiae) |
| chr13_-_22042949 | 4.73 |
ENSMUST00000091741.4
|
Hist1h2ag
|
histone cluster 1, H2ag |
| chr13_+_21810428 | 4.72 |
ENSMUST00000091745.5
|
Hist1h2ao
|
histone cluster 1, H2ao |
| chr2_+_72476225 | 4.63 |
ENSMUST00000157019.1
|
Cdca7
|
cell division cycle associated 7 |
| chr13_+_22043189 | 4.62 |
ENSMUST00000110452.1
|
Hist1h2bj
|
histone cluster 1, H2bj |
| chr18_+_56707725 | 4.41 |
ENSMUST00000025486.8
|
Lmnb1
|
lamin B1 |
| chr10_+_11281304 | 4.34 |
ENSMUST00000129456.1
|
Fbxo30
|
F-box protein 30 |
| chr10_+_11281583 | 4.33 |
ENSMUST00000070300.4
|
Fbxo30
|
F-box protein 30 |
| chr7_-_44548733 | 3.96 |
ENSMUST00000145956.1
ENSMUST00000049343.8 |
Pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr12_+_73286868 | 3.94 |
ENSMUST00000153941.1
ENSMUST00000122920.1 ENSMUST00000101313.3 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr13_-_21783391 | 3.78 |
ENSMUST00000099704.3
|
Hist1h3i
|
histone cluster 1, H3i |
| chr11_-_69560186 | 3.70 |
ENSMUST00000004036.5
|
Efnb3
|
ephrin B3 |
| chr14_-_20388822 | 3.70 |
ENSMUST00000022345.6
|
Dnajc9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| chr12_-_11265768 | 3.58 |
ENSMUST00000166117.1
|
Gen1
|
Gen homolog 1, endonuclease (Drosophila) |
| chr9_+_3017408 | 3.47 |
ENSMUST00000099049.3
|
Gm10719
|
predicted gene 10719 |
| chr6_-_67037399 | 3.43 |
ENSMUST00000043098.6
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr13_-_55329723 | 3.39 |
ENSMUST00000021941.7
|
Mxd3
|
Max dimerization protein 3 |
| chr1_-_128359610 | 3.26 |
ENSMUST00000027601.4
|
Mcm6
|
minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) |
| chr17_-_35516780 | 3.11 |
ENSMUST00000160885.1
ENSMUST00000159009.1 ENSMUST00000161012.1 |
Tcf19
|
transcription factor 19 |
| chr7_+_66109474 | 3.04 |
ENSMUST00000036372.6
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chr13_+_23581563 | 2.89 |
ENSMUST00000102968.1
|
Hist1h4d
|
histone cluster 1, H4d |
| chr3_+_88532314 | 2.81 |
ENSMUST00000172699.1
|
Mex3a
|
mex3 homolog A (C. elegans) |
| chr4_+_132768325 | 2.79 |
ENSMUST00000102561.4
|
Rpa2
|
replication protein A2 |
| chr1_-_180813591 | 2.76 |
ENSMUST00000162118.1
ENSMUST00000159685.1 ENSMUST00000161308.1 |
H3f3a
|
H3 histone, family 3A |
| chr11_-_87404380 | 2.69 |
ENSMUST00000067692.6
|
Rad51c
|
RAD51 homolog C |
| chr9_+_64281575 | 2.61 |
ENSMUST00000034964.6
|
Tipin
|
timeless interacting protein |
| chr9_+_3015654 | 2.60 |
ENSMUST00000099050.3
|
Gm10720
|
predicted gene 10720 |
| chr13_+_21716385 | 2.54 |
ENSMUST00000070124.3
|
Hist1h2ai
|
histone cluster 1, H2ai |
| chr13_+_23555023 | 2.48 |
ENSMUST00000045301.6
|
Hist1h1d
|
histone cluster 1, H1d |
| chr1_-_156474249 | 2.46 |
ENSMUST00000051396.6
|
Soat1
|
sterol O-acyltransferase 1 |
| chr13_-_22035589 | 2.43 |
ENSMUST00000091742.4
|
Hist1h2ah
|
histone cluster 1, H2ah |
| chr9_+_3013140 | 2.43 |
ENSMUST00000143083.2
|
Gm10721
|
predicted gene 10721 |
| chr11_-_70410010 | 2.39 |
ENSMUST00000019065.3
ENSMUST00000135148.1 |
Pelp1
|
proline, glutamic acid and leucine rich protein 1 |
| chr10_-_35711891 | 2.22 |
ENSMUST00000080898.2
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chrX_+_153006461 | 2.20 |
ENSMUST00000095755.3
|
Usp51
|
ubiquitin specific protease 51 |
| chr14_+_79451791 | 2.17 |
ENSMUST00000100359.1
|
Zbtbd6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr7_+_35802593 | 2.14 |
ENSMUST00000052454.2
|
E130304I02Rik
|
RIKEN cDNA E130304I02 gene |
| chr9_+_3025417 | 2.10 |
ENSMUST00000075573.6
|
Gm10717
|
predicted gene 10717 |
| chr13_+_23574381 | 2.09 |
ENSMUST00000090776.4
|
Hist1h2ad
|
histone cluster 1, H2ad |
| chr13_+_109685994 | 2.04 |
ENSMUST00000074103.5
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chrX_-_37085402 | 1.99 |
ENSMUST00000115231.3
|
Rpl39
|
ribosomal protein L39 |
| chr16_+_38346986 | 1.99 |
ENSMUST00000050273.8
ENSMUST00000120495.1 ENSMUST00000119704.1 |
Cox17
Gm21987
|
cytochrome c oxidase assembly protein 17 predicted gene 21987 |
| chr18_+_44104407 | 1.98 |
ENSMUST00000081271.5
|
Spink12
|
serine peptidase inhibitor, Kazal type 11 |
| chr1_+_172521044 | 1.97 |
ENSMUST00000085894.5
ENSMUST00000161140.1 ENSMUST00000162988.1 |
Ccdc19
|
coiled-coil domain containing 19 |
| chr7_+_110122299 | 1.96 |
ENSMUST00000033326.8
|
Wee1
|
WEE 1 homolog 1 (S. pombe) |
| chr15_+_82298943 | 1.81 |
ENSMUST00000023089.3
|
Wbp2nl
|
WBP2 N-terminal like |
| chr1_+_74506044 | 1.79 |
ENSMUST00000087215.5
|
Rqcd1
|
rcd1 (required for cell differentiation) homolog 1 (S. pombe) |
| chr1_+_136683375 | 1.76 |
ENSMUST00000181524.1
|
Gm19705
|
predicted gene, 19705 |
| chr9_+_3037111 | 1.74 |
ENSMUST00000177969.1
|
Gm10715
|
predicted gene 10715 |
| chr5_+_129020069 | 1.73 |
ENSMUST00000031383.7
ENSMUST00000111343.1 |
Ran
|
RAN, member RAS oncogene family |
| chr4_+_124714776 | 1.71 |
ENSMUST00000030734.4
|
Sf3a3
|
splicing factor 3a, subunit 3 |
| chr10_-_117376922 | 1.55 |
ENSMUST00000177145.1
ENSMUST00000176670.1 |
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr13_+_21754067 | 1.43 |
ENSMUST00000091709.2
|
Hist1h2bn
|
histone cluster 1, H2bn |
| chr5_+_135187251 | 1.43 |
ENSMUST00000002825.5
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr9_+_44334685 | 1.41 |
ENSMUST00000052686.2
|
H2afx
|
H2A histone family, member X |
| chr15_+_84720052 | 1.38 |
ENSMUST00000006029.4
ENSMUST00000172307.2 |
Arhgap8
|
Rho GTPase activating protein 8 |
| chr19_+_8723478 | 1.38 |
ENSMUST00000180819.1
ENSMUST00000181422.1 |
Snhg1
|
small nucleolar RNA host gene (non-protein coding) 1 |
| chrX_+_68678541 | 1.34 |
ENSMUST00000088546.5
|
Fmr1
|
fragile X mental retardation syndrome 1 |
| chr3_+_14533817 | 1.29 |
ENSMUST00000169079.1
ENSMUST00000091325.3 |
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr12_-_12941827 | 1.29 |
ENSMUST00000043396.7
|
Mycn
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) |
| chr7_-_28598140 | 1.27 |
ENSMUST00000040531.8
ENSMUST00000108283.1 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr7_-_45896656 | 1.26 |
ENSMUST00000120299.1
|
Syngr4
|
synaptogyrin 4 |
| chr17_+_34850373 | 1.25 |
ENSMUST00000097343.4
ENSMUST00000173357.1 ENSMUST00000173065.1 ENSMUST00000165953.2 |
Nelfe
|
negative elongation factor complex member E, Rdbp |
| chr2_-_157204483 | 1.14 |
ENSMUST00000029170.7
|
Rbl1
|
retinoblastoma-like 1 (p107) |
| chr11_-_106301801 | 1.13 |
ENSMUST00000103071.3
|
Gh
|
growth hormone |
| chr1_+_118389058 | 1.10 |
ENSMUST00000049404.6
ENSMUST00000070989.7 ENSMUST00000165223.1 ENSMUST00000178710.1 |
Clasp1
|
CLIP associating protein 1 |
| chr7_+_28169744 | 1.08 |
ENSMUST00000042405.6
|
Fbl
|
fibrillarin |
| chr10_-_117376955 | 1.05 |
ENSMUST00000069168.6
ENSMUST00000176686.1 |
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr11_+_43681998 | 1.04 |
ENSMUST00000061070.5
|
Pwwp2a
|
PWWP domain containing 2A |
| chr4_-_108579330 | 1.04 |
ENSMUST00000079213.5
|
Prpf38a
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing A |
| chr11_+_43682038 | 1.03 |
ENSMUST00000094294.4
|
Pwwp2a
|
PWWP domain containing 2A |
| chr3_+_14533788 | 1.00 |
ENSMUST00000108370.2
|
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr11_+_87127267 | 0.89 |
ENSMUST00000139532.1
|
Trim37
|
tripartite motif-containing 37 |
| chr15_-_81871883 | 0.88 |
ENSMUST00000023117.8
|
Phf5a
|
PHD finger protein 5A |
| chr7_+_44816088 | 0.87 |
ENSMUST00000057195.9
ENSMUST00000107891.1 |
Nup62
|
nucleoporin 62 |
| chrX_-_73966329 | 0.87 |
ENSMUST00000114372.2
ENSMUST00000033761.6 |
Hcfc1
|
host cell factor C1 |
| chr1_-_191183244 | 0.86 |
ENSMUST00000027941.8
|
Atf3
|
activating transcription factor 3 |
| chr13_+_22035821 | 0.86 |
ENSMUST00000110455.2
|
Hist1h2bk
|
histone cluster 1, H2bk |
| chr2_+_179442427 | 0.85 |
ENSMUST00000000314.6
|
Cdh4
|
cadherin 4 |
| chr17_-_35046726 | 0.84 |
ENSMUST00000097338.4
|
Msh5
|
mutS homolog 5 (E. coli) |
| chr3_-_57847478 | 0.79 |
ENSMUST00000120289.1
ENSMUST00000066882.8 |
Pfn2
|
profilin 2 |
| chr12_+_55836365 | 0.78 |
ENSMUST00000059250.6
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr3_+_14533867 | 0.77 |
ENSMUST00000163660.1
|
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr4_+_136286061 | 0.75 |
ENSMUST00000069195.4
ENSMUST00000130658.1 |
Zfp46
|
zinc finger protein 46 |
| chr11_-_77513335 | 0.73 |
ENSMUST00000060417.4
|
Trp53i13
|
transformation related protein 53 inducible protein 13 |
| chr5_-_137533170 | 0.70 |
ENSMUST00000168746.1
ENSMUST00000170293.1 |
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr7_-_45694369 | 0.69 |
ENSMUST00000040636.6
|
Sec1
|
secretory blood group 1 |
| chr4_+_94739276 | 0.62 |
ENSMUST00000073939.6
ENSMUST00000102798.1 |
Tek
|
endothelial-specific receptor tyrosine kinase |
| chr9_-_121277160 | 0.59 |
ENSMUST00000051479.6
ENSMUST00000171923.1 |
Ulk4
|
unc-51-like kinase 4 |
| chr17_+_12916329 | 0.50 |
ENSMUST00000089024.6
ENSMUST00000151287.1 ENSMUST00000143961.1 |
Tcp1
|
t-complex protein 1 |
| chr13_-_21810190 | 0.49 |
ENSMUST00000110469.1
ENSMUST00000091749.2 |
Hist1h2bq
|
histone cluster 1, H2bq |
| chr12_+_112146187 | 0.48 |
ENSMUST00000128402.2
|
Kif26a
|
kinesin family member 26A |
| chrX_+_162760388 | 0.47 |
ENSMUST00000033720.5
ENSMUST00000112327.1 |
Rbbp7
|
retinoblastoma binding protein 7 |
| chr11_+_85311232 | 0.45 |
ENSMUST00000020835.9
|
Ppm1d
|
protein phosphatase 1D magnesium-dependent, delta isoform |
| chrX_+_68678624 | 0.43 |
ENSMUST00000114656.1
|
Fmr1
|
fragile X mental retardation syndrome 1 |
| chr2_+_147364989 | 0.42 |
ENSMUST00000109968.2
|
Pax1
|
paired box gene 1 |
| chr19_+_34550664 | 0.38 |
ENSMUST00000149829.1
ENSMUST00000102826.3 |
Ifit2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr17_-_34850162 | 0.37 |
ENSMUST00000046022.9
|
Skiv2l
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr11_+_88047693 | 0.34 |
ENSMUST00000079866.4
|
Srsf1
|
serine/arginine-rich splicing factor 1 |
| chr1_+_180568913 | 0.34 |
ENSMUST00000027777.6
|
Parp1
|
poly (ADP-ribose) polymerase family, member 1 |
| chr13_-_21716143 | 0.30 |
ENSMUST00000091756.1
|
Hist1h2bl
|
histone cluster 1, H2bl |
| chrX_+_71981522 | 0.29 |
ENSMUST00000114569.1
|
Fate1
|
fetal and adult testis expressed 1 |
| chr6_-_52183828 | 0.28 |
ENSMUST00000134831.1
|
Hoxa3
|
homeobox A3 |
| chr6_-_30509706 | 0.24 |
ENSMUST00000064330.6
ENSMUST00000102991.2 ENSMUST00000115157.1 ENSMUST00000148638.1 |
Tmem209
|
transmembrane protein 209 |
| chr15_-_98093245 | 0.22 |
ENSMUST00000180657.1
|
Senp1
|
SUMO1/sentrin specific peptidase 1 |
| chr13_-_97198351 | 0.17 |
ENSMUST00000022169.7
|
Hexb
|
hexosaminidase B |
| chr3_+_60501252 | 0.16 |
ENSMUST00000099087.2
|
Mbnl1
|
muscleblind-like 1 (Drosophila) |
| chr17_+_35517100 | 0.13 |
ENSMUST00000164242.2
ENSMUST00000045956.7 |
Cchcr1
|
coiled-coil alpha-helical rod protein 1 |
| chr5_-_92348871 | 0.10 |
ENSMUST00000038816.6
ENSMUST00000118006.1 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr17_+_33524170 | 0.09 |
ENSMUST00000087623.6
|
Adamts10
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 10 |
| chr6_+_113077631 | 0.09 |
ENSMUST00000113157.1
|
Setd5
|
SET domain containing 5 |
| chr9_+_111271832 | 0.08 |
ENSMUST00000060711.5
|
Epm2aip1
|
EPM2A (laforin) interacting protein 1 |
| chr2_+_11172080 | 0.02 |
ENSMUST00000114853.1
|
Prkcq
|
protein kinase C, theta |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 15.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 1.5 | 4.4 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 1.4 | 8.7 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) tetrahydrofolate biosynthetic process(GO:0046654) |
| 1.3 | 4.0 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 1.1 | 7.4 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 1.0 | 11.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.9 | 16.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.9 | 11.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.8 | 8.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.7 | 2.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.7 | 2.8 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.7 | 10.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.7 | 6.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.7 | 2.7 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.7 | 2.0 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.6 | 1.8 | GO:1901254 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.5 | 2.6 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.4 | 3.7 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.4 | 3.6 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.3 | 5.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.3 | 2.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 3.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.3 | 0.9 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.3 | 0.9 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.3 | 3.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 2.5 | GO:0042984 | very-low-density lipoprotein particle assembly(GO:0034379) amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 5.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 24.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.2 | 1.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 0.9 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.2 | 6.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.2 | 1.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 6.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.2 | 0.9 | GO:0046600 | histone H2A-K119 monoubiquitination(GO:0036353) negative regulation of centriole replication(GO:0046600) |
| 0.2 | 2.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 2.0 | GO:1901898 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.8 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 1.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 2.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.5 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.5 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.1 | 0.3 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.1 | 1.3 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 3.1 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.5 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 3.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.8 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 1.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 2.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.4 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.4 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 4.5 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 2.6 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 6.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 1.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 1.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.4 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 2.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.6 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 1.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 2.0 | GO:0030010 | establishment of cell polarity(GO:0030010) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 16.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 2.5 | 20.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 1.3 | 4.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.6 | 4.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.6 | 1.7 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.5 | 2.7 | GO:0048476 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) Holliday junction resolvase complex(GO:0048476) |
| 0.4 | 1.8 | GO:1902737 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 0.4 | 2.8 | GO:0001740 | Barr body(GO:0001740) |
| 0.4 | 5.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.4 | 2.6 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.4 | 0.4 | GO:0055087 | Ski complex(GO:0055087) |
| 0.3 | 2.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 1.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.3 | 20.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.3 | 1.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 13.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 1.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.2 | 2.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 3.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 10.5 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 11.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 2.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.4 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 0.9 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.1 | 1.9 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 10.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 5.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 13.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.5 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 16.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 2.2 | 8.7 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 2.2 | 10.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.9 | 11.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.0 | 3.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.8 | 6.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.8 | 10.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 5.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 7.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.3 | 2.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.3 | 1.7 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.3 | 4.0 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.3 | 1.8 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.2 | 2.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 0.7 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.2 | 4.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.9 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 1.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 6.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 10.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.9 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 3.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 3.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 2.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 10.1 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 6.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 9.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 3.7 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 3.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 6.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 3.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 6.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.1 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 9.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 7.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.5 | 21.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.4 | 32.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 2.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 5.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 8.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 4.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.4 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 22.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 1.5 | 7.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 1.2 | 16.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 1.1 | 14.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.8 | 6.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.4 | 21.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 6.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 2.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 6.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 2.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 3.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 3.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 4.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 1.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 2.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 2.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |