Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
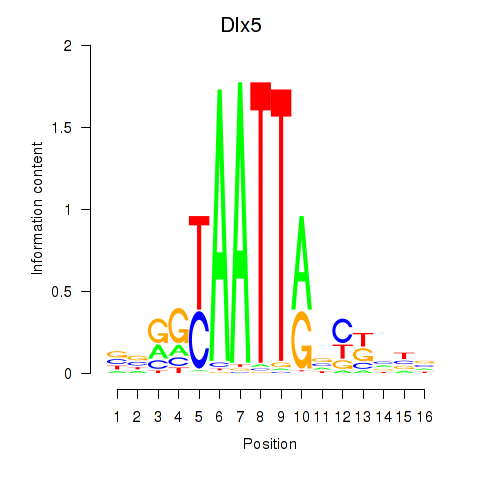
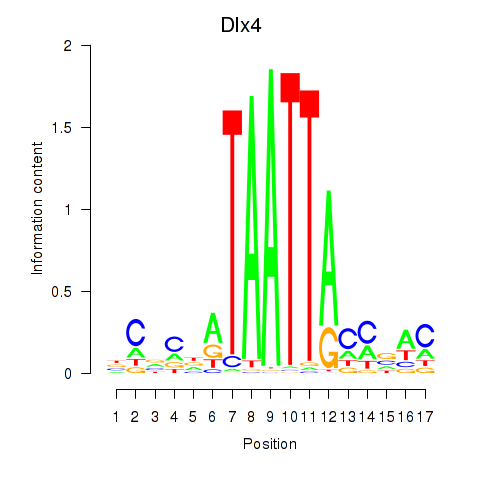
Results for Dlx5_Dlx4
Z-value: 0.79
Transcription factors associated with Dlx5_Dlx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dlx5
|
ENSMUSG00000029755.9 | distal-less homeobox 5 |
|
Dlx4
|
ENSMUSG00000020871.7 | distal-less homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx5 | mm10_v2_chr6_-_6882068_6882092 | 0.24 | 1.5e-01 | Click! |
| Dlx4 | mm10_v2_chr11_-_95146263_95146263 | -0.16 | 3.7e-01 | Click! |
Activity profile of Dlx5_Dlx4 motif
Sorted Z-values of Dlx5_Dlx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_5799491 | 2.66 |
ENSMUST00000181484.1
|
3300005D01Rik
|
RIKEN cDNA 3300005D01 gene |
| chr10_-_76110956 | 2.44 |
ENSMUST00000120757.1
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr16_-_75909272 | 1.96 |
ENSMUST00000114239.2
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr8_-_4779513 | 1.91 |
ENSMUST00000022945.7
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr19_-_32196393 | 1.84 |
ENSMUST00000151822.1
|
Sgms1
|
sphingomyelin synthase 1 |
| chr12_-_55014329 | 1.78 |
ENSMUST00000172875.1
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr19_-_40588374 | 1.67 |
ENSMUST00000175932.1
ENSMUST00000176955.1 ENSMUST00000149476.2 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr17_-_37280418 | 1.63 |
ENSMUST00000077585.2
|
Olfr99
|
olfactory receptor 99 |
| chr7_+_104003259 | 1.61 |
ENSMUST00000098184.1
|
Olfr638
|
olfactory receptor 638 |
| chr16_+_36277145 | 1.59 |
ENSMUST00000042097.9
|
Stfa1
|
stefin A1 |
| chr4_-_132075250 | 1.54 |
ENSMUST00000105970.1
ENSMUST00000105975.1 |
Epb4.1
|
erythrocyte protein band 4.1 |
| chr12_-_110696248 | 1.39 |
ENSMUST00000124156.1
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr9_-_15357692 | 1.38 |
ENSMUST00000098979.3
ENSMUST00000161132.1 |
5830418K08Rik
|
RIKEN cDNA 5830418K08 gene |
| chr2_-_126500631 | 1.32 |
ENSMUST00000129187.1
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr15_-_101694299 | 1.28 |
ENSMUST00000023788.6
|
Krt6a
|
keratin 6A |
| chr1_+_107535508 | 1.26 |
ENSMUST00000182198.1
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr8_-_85380964 | 1.20 |
ENSMUST00000122452.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr11_+_31832660 | 1.19 |
ENSMUST00000132857.1
|
Gm12107
|
predicted gene 12107 |
| chr9_+_95857597 | 1.18 |
ENSMUST00000034980.7
|
Atr
|
ataxia telangiectasia and Rad3 related |
| chr3_+_9403049 | 1.17 |
ENSMUST00000180874.1
ENSMUST00000181331.1 ENSMUST00000181930.1 |
C030034L19Rik
|
RIKEN cDNA C030034L19 gene |
| chr16_+_45610380 | 1.16 |
ENSMUST00000161347.2
ENSMUST00000023339.4 |
Gcsam
|
germinal center associated, signaling and motility |
| chr17_+_40811089 | 1.15 |
ENSMUST00000024721.7
|
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr19_+_55180799 | 1.13 |
ENSMUST00000025936.5
|
Tectb
|
tectorin beta |
| chr19_+_23723279 | 1.13 |
ENSMUST00000067077.1
|
Gm9938
|
predicted gene 9938 |
| chr13_+_23746734 | 1.13 |
ENSMUST00000099703.2
|
Hist1h2bb
|
histone cluster 1, H2bb |
| chr11_+_87793470 | 1.12 |
ENSMUST00000020779.4
|
Mpo
|
myeloperoxidase |
| chr8_-_85365317 | 1.11 |
ENSMUST00000034133.7
|
Mylk3
|
myosin light chain kinase 3 |
| chr2_-_121235689 | 1.10 |
ENSMUST00000142400.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr2_+_131491764 | 1.10 |
ENSMUST00000028806.5
ENSMUST00000110179.2 ENSMUST00000110189.2 ENSMUST00000110182.2 ENSMUST00000110183.2 ENSMUST00000110186.2 ENSMUST00000110188.1 |
Smox
|
spermine oxidase |
| chr11_-_17008647 | 1.09 |
ENSMUST00000102881.3
|
Plek
|
pleckstrin |
| chr15_+_25773985 | 1.08 |
ENSMUST00000125667.1
|
Myo10
|
myosin X |
| chr5_+_90768511 | 1.06 |
ENSMUST00000031319.6
|
Ppbp
|
pro-platelet basic protein |
| chr9_-_95750335 | 1.05 |
ENSMUST00000053785.3
|
Trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr4_-_123664725 | 1.03 |
ENSMUST00000147030.1
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chrX_-_134111852 | 1.02 |
ENSMUST00000033610.6
|
Nox1
|
NADPH oxidase 1 |
| chrX_+_159303266 | 1.02 |
ENSMUST00000112491.1
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chrX_+_93675088 | 1.01 |
ENSMUST00000045898.3
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr13_+_104229366 | 1.00 |
ENSMUST00000022227.6
|
Cenpk
|
centromere protein K |
| chr13_+_23555023 | 1.00 |
ENSMUST00000045301.6
|
Hist1h1d
|
histone cluster 1, H1d |
| chr3_-_116253467 | 0.99 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr6_+_5390387 | 0.98 |
ENSMUST00000183358.1
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr19_-_40588338 | 0.97 |
ENSMUST00000176939.1
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr14_+_32321987 | 0.97 |
ENSMUST00000022480.7
|
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chrX_-_7978027 | 0.96 |
ENSMUST00000125418.1
|
Gata1
|
GATA binding protein 1 |
| chr1_+_139454747 | 0.94 |
ENSMUST00000053364.8
ENSMUST00000097554.3 |
Aspm
|
asp (abnormal spindle)-like, microcephaly associated (Drosophila) |
| chr14_+_27000362 | 0.93 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr6_-_67376147 | 0.92 |
ENSMUST00000018485.3
|
Il12rb2
|
interleukin 12 receptor, beta 2 |
| chr9_-_36726374 | 0.92 |
ENSMUST00000172702.2
ENSMUST00000172742.1 ENSMUST00000034625.5 |
Chek1
|
checkpoint kinase 1 |
| chr7_-_133702515 | 0.92 |
ENSMUST00000153698.1
|
Uros
|
uroporphyrinogen III synthase |
| chr17_+_21690766 | 0.89 |
ENSMUST00000097384.1
|
Gm10509
|
predicted gene 10509 |
| chr8_-_122915987 | 0.87 |
ENSMUST00000098333.4
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr8_-_22060019 | 0.86 |
ENSMUST00000110738.2
|
Atp7b
|
ATPase, Cu++ transporting, beta polypeptide |
| chr5_-_62765618 | 0.86 |
ENSMUST00000159470.1
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr8_+_21391811 | 0.85 |
ENSMUST00000120874.3
|
Gm21002
|
predicted gene, 21002 |
| chr3_-_153792087 | 0.85 |
ENSMUST00000081193.1
|
5730460C07Rik
|
RIKEN cDNA 5730460C07 gene |
| chr18_+_4993795 | 0.83 |
ENSMUST00000153016.1
|
Svil
|
supervillin |
| chr1_+_58210397 | 0.83 |
ENSMUST00000040442.5
|
Aox4
|
aldehyde oxidase 4 |
| chr17_+_3532554 | 0.83 |
ENSMUST00000168560.1
|
Cldn20
|
claudin 20 |
| chr1_+_40439627 | 0.82 |
ENSMUST00000097772.3
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr5_+_76588663 | 0.82 |
ENSMUST00000121979.1
|
Cep135
|
centrosomal protein 135 |
| chr16_+_92478743 | 0.81 |
ENSMUST00000160494.1
|
2410124H12Rik
|
RIKEN cDNA 2410124H12 gene |
| chrX_+_56454871 | 0.81 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr7_+_16781341 | 0.80 |
ENSMUST00000108496.2
|
Slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr14_+_79515618 | 0.80 |
ENSMUST00000110835.1
|
Elf1
|
E74-like factor 1 |
| chr4_+_126556994 | 0.80 |
ENSMUST00000147675.1
|
Clspn
|
claspin |
| chrX_-_53240101 | 0.78 |
ENSMUST00000074861.2
|
Plac1
|
placental specific protein 1 |
| chr9_+_65890237 | 0.77 |
ENSMUST00000045802.6
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene |
| chr13_+_23575753 | 0.77 |
ENSMUST00000105105.1
|
Hist1h3d
|
histone cluster 1, H3d |
| chr2_-_37703845 | 0.77 |
ENSMUST00000155237.1
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chrX_+_56787701 | 0.77 |
ENSMUST00000151033.1
|
Fhl1
|
four and a half LIM domains 1 |
| chr16_-_17144415 | 0.75 |
ENSMUST00000115709.1
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr10_+_75564086 | 0.75 |
ENSMUST00000141062.1
ENSMUST00000152657.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr13_-_95478655 | 0.75 |
ENSMUST00000022186.3
|
S100z
|
S100 calcium binding protein, zeta |
| chr1_-_174250976 | 0.74 |
ENSMUST00000061990.4
|
Olfr419
|
olfactory receptor 419 |
| chr2_-_140066661 | 0.74 |
ENSMUST00000046656.2
ENSMUST00000099304.3 ENSMUST00000110079.2 |
Tasp1
|
taspase, threonine aspartase 1 |
| chr9_-_53248106 | 0.74 |
ENSMUST00000065630.6
|
Ddx10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr13_+_21717626 | 0.72 |
ENSMUST00000091754.2
|
Hist1h3h
|
histone cluster 1, H3h |
| chr7_-_143460989 | 0.72 |
ENSMUST00000167912.1
ENSMUST00000037287.6 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C (P57) |
| chr1_+_40439767 | 0.71 |
ENSMUST00000173514.1
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr16_+_94425083 | 0.71 |
ENSMUST00000141176.1
|
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr1_-_126738167 | 0.70 |
ENSMUST00000160693.1
|
Nckap5
|
NCK-associated protein 5 |
| chr13_+_22327911 | 0.69 |
ENSMUST00000091734.1
|
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chr6_-_50456085 | 0.69 |
ENSMUST00000146341.1
ENSMUST00000071728.4 |
Osbpl3
|
oxysterol binding protein-like 3 |
| chr9_-_20959785 | 0.69 |
ENSMUST00000177754.1
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr8_+_21287409 | 0.69 |
ENSMUST00000098893.3
|
Defa3
|
defensin, alpha, 3 |
| chr4_+_19818722 | 0.68 |
ENSMUST00000035890.7
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr4_-_154928545 | 0.68 |
ENSMUST00000152687.1
|
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chrX_+_71962971 | 0.68 |
ENSMUST00000048790.6
|
Prrg3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr16_+_36253046 | 0.68 |
ENSMUST00000063539.6
|
2010005H15Rik
|
RIKEN cDNA 2010005H15 gene |
| chr4_-_41464816 | 0.68 |
ENSMUST00000108055.2
ENSMUST00000154535.1 ENSMUST00000030148.5 |
Kif24
|
kinesin family member 24 |
| chr2_+_121956651 | 0.67 |
ENSMUST00000110574.1
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr10_+_26822560 | 0.67 |
ENSMUST00000135866.1
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr11_+_58948890 | 0.67 |
ENSMUST00000078267.3
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr5_-_73191848 | 0.67 |
ENSMUST00000176910.1
|
Fryl
|
furry homolog-like (Drosophila) |
| chr18_-_43477764 | 0.67 |
ENSMUST00000057110.9
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr7_+_29983948 | 0.67 |
ENSMUST00000148442.1
|
Zfp568
|
zinc finger protein 568 |
| chr1_-_65123108 | 0.66 |
ENSMUST00000050047.3
ENSMUST00000148020.1 |
D630023F18Rik
|
RIKEN cDNA D630023F18 gene |
| chr13_+_44729535 | 0.65 |
ENSMUST00000174068.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr5_-_148371525 | 0.65 |
ENSMUST00000138596.1
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr1_-_134955847 | 0.64 |
ENSMUST00000168381.1
|
Ppp1r12b
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B |
| chr4_+_126556935 | 0.64 |
ENSMUST00000048391.8
|
Clspn
|
claspin |
| chrX_-_160906998 | 0.64 |
ENSMUST00000069417.5
|
Gja6
|
gap junction protein, alpha 6 |
| chr11_+_73350839 | 0.64 |
ENSMUST00000120137.1
|
Olfr20
|
olfactory receptor 20 |
| chr3_+_88629442 | 0.63 |
ENSMUST00000176316.1
ENSMUST00000176879.1 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr2_+_176711933 | 0.63 |
ENSMUST00000108983.2
|
Gm14305
|
predicted gene 14305 |
| chr8_+_21191614 | 0.63 |
ENSMUST00000098896.4
|
Defa-rs7
|
defensin, alpha, related sequence 7 |
| chr10_-_117148474 | 0.62 |
ENSMUST00000020381.3
|
Frs2
|
fibroblast growth factor receptor substrate 2 |
| chr3_+_79884576 | 0.60 |
ENSMUST00000145992.1
|
Fam198b
|
family with sequence similarity 198, member B |
| chr8_+_21134610 | 0.60 |
ENSMUST00000098898.4
|
Gm15284
|
predicted gene 15284 |
| chr2_+_119047116 | 0.60 |
ENSMUST00000152380.1
ENSMUST00000099542.2 |
Casc5
|
cancer susceptibility candidate 5 |
| chrM_+_11734 | 0.60 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr17_-_26099257 | 0.60 |
ENSMUST00000053575.3
|
Gm8186
|
predicted gene 8186 |
| chrX_-_112405822 | 0.59 |
ENSMUST00000124335.1
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr2_+_118814195 | 0.59 |
ENSMUST00000110842.1
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr6_+_92092369 | 0.59 |
ENSMUST00000113463.1
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr15_-_34356421 | 0.59 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr11_+_75486813 | 0.58 |
ENSMUST00000018449.4
ENSMUST00000102510.1 ENSMUST00000131283.1 |
Prpf8
|
pre-mRNA processing factor 8 |
| chr3_+_88629499 | 0.57 |
ENSMUST00000175745.1
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr6_-_66559708 | 0.57 |
ENSMUST00000079584.1
|
Vmn1r32
|
vomeronasal 1 receptor 32 |
| chr8_+_23411490 | 0.57 |
ENSMUST00000033952.7
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr2_+_118813995 | 0.57 |
ENSMUST00000134661.1
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr2_+_4300462 | 0.56 |
ENSMUST00000175669.1
|
Frmd4a
|
FERM domain containing 4A |
| chr8_+_116921735 | 0.56 |
ENSMUST00000034205.4
|
Cenpn
|
centromere protein N |
| chr13_-_89742244 | 0.56 |
ENSMUST00000109543.2
ENSMUST00000159337.1 ENSMUST00000159910.1 ENSMUST00000109544.2 |
Vcan
|
versican |
| chr11_+_46454921 | 0.54 |
ENSMUST00000020668.8
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr8_+_95055094 | 0.54 |
ENSMUST00000058479.6
|
Ccdc135
|
coiled-coil domain containing 135 |
| chr12_-_110695860 | 0.54 |
ENSMUST00000149189.1
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr14_-_104522615 | 0.54 |
ENSMUST00000022716.2
|
Rnf219
|
ring finger protein 219 |
| chr13_-_62371936 | 0.53 |
ENSMUST00000107989.3
|
Gm3604
|
predicted gene 3604 |
| chr9_+_113812547 | 0.53 |
ENSMUST00000166734.2
ENSMUST00000111838.2 ENSMUST00000163895.2 |
Clasp2
|
CLIP associating protein 2 |
| chr2_-_168767136 | 0.53 |
ENSMUST00000029061.5
ENSMUST00000103074.1 |
Sall4
|
sal-like 4 (Drosophila) |
| chrX_+_150547375 | 0.53 |
ENSMUST00000066337.6
ENSMUST00000112715.1 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr15_+_84232030 | 0.53 |
ENSMUST00000023072.6
|
Parvb
|
parvin, beta |
| chr2_+_118814237 | 0.52 |
ENSMUST00000028803.7
ENSMUST00000126045.1 |
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr12_-_101028983 | 0.52 |
ENSMUST00000068411.3
ENSMUST00000085096.3 |
Ccdc88c
|
coiled-coil domain containing 88C |
| chr3_-_98339921 | 0.52 |
ENSMUST00000065793.5
|
Phgdh
|
3-phosphoglycerate dehydrogenase |
| chr12_+_116275386 | 0.52 |
ENSMUST00000090195.4
|
Gm11027
|
predicted gene 11027 |
| chr13_+_93308006 | 0.52 |
ENSMUST00000079086.6
|
Homer1
|
homer homolog 1 (Drosophila) |
| chr19_-_56548013 | 0.51 |
ENSMUST00000182059.1
|
Dclre1a
|
DNA cross-link repair 1A, PSO2 homolog (S. cerevisiae) |
| chr8_+_105297663 | 0.51 |
ENSMUST00000015003.8
|
E2f4
|
E2F transcription factor 4 |
| chr3_+_59952185 | 0.51 |
ENSMUST00000094227.3
|
Gm9696
|
predicted gene 9696 |
| chr6_-_129275360 | 0.50 |
ENSMUST00000032259.3
|
Cd69
|
CD69 antigen |
| chr3_+_146121655 | 0.49 |
ENSMUST00000039450.4
|
Mcoln3
|
mucolipin 3 |
| chr4_+_8690399 | 0.49 |
ENSMUST00000127476.1
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr12_+_55124528 | 0.49 |
ENSMUST00000177768.1
|
Fam177a
|
family with sequence similarity 177, member A |
| chr10_-_62814539 | 0.49 |
ENSMUST00000173087.1
ENSMUST00000174121.1 |
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr12_+_38780284 | 0.48 |
ENSMUST00000162563.1
ENSMUST00000161164.1 ENSMUST00000160996.1 |
Etv1
|
ets variant gene 1 |
| chr2_+_121956411 | 0.48 |
ENSMUST00000110578.1
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chrX_+_56786527 | 0.48 |
ENSMUST00000144600.1
|
Fhl1
|
four and a half LIM domains 1 |
| chrX_+_101794644 | 0.48 |
ENSMUST00000151231.2
|
8030474K03Rik
|
RIKEN cDNA 8030474K03 gene |
| chr4_+_49631999 | 0.47 |
ENSMUST00000140341.1
|
Rnf20
|
ring finger protein 20 |
| chr11_+_87109221 | 0.47 |
ENSMUST00000020794.5
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr13_+_21716385 | 0.47 |
ENSMUST00000070124.3
|
Hist1h2ai
|
histone cluster 1, H2ai |
| chr6_-_120357440 | 0.47 |
ENSMUST00000112703.1
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr6_+_72097561 | 0.47 |
ENSMUST00000069994.4
ENSMUST00000114112.1 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr4_+_133176336 | 0.47 |
ENSMUST00000105912.1
|
Wasf2
|
WAS protein family, member 2 |
| chr4_-_42034726 | 0.47 |
ENSMUST00000084677.2
|
Gm21093
|
predicted gene, 21093 |
| chr10_+_80264942 | 0.47 |
ENSMUST00000105362.1
ENSMUST00000105361.3 |
Dazap1
|
DAZ associated protein 1 |
| chr2_-_45110336 | 0.47 |
ENSMUST00000028229.6
ENSMUST00000152232.1 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_20900867 | 0.46 |
ENSMUST00000079237.5
|
Zfp125
|
zinc finger protein 125 |
| chr14_+_53806497 | 0.46 |
ENSMUST00000103672.4
|
Trav17
|
T cell receptor alpha variable 17 |
| chr4_-_119658781 | 0.46 |
ENSMUST00000106309.2
ENSMUST00000044426.7 |
Guca2b
|
guanylate cyclase activator 2b (retina) |
| chr13_+_94083490 | 0.46 |
ENSMUST00000156071.1
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr2_-_89024099 | 0.46 |
ENSMUST00000099799.2
|
Olfr1217
|
olfactory receptor 1217 |
| chr19_-_8819278 | 0.46 |
ENSMUST00000088092.5
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr13_-_12258093 | 0.45 |
ENSMUST00000099856.4
|
Mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr16_+_34690548 | 0.45 |
ENSMUST00000023532.6
|
Ccdc14
|
coiled-coil domain containing 14 |
| chr2_+_174450678 | 0.45 |
ENSMUST00000016399.5
|
Tubb1
|
tubulin, beta 1 class VI |
| chr19_+_55180716 | 0.45 |
ENSMUST00000154886.1
ENSMUST00000120936.1 |
Tectb
|
tectorin beta |
| chr17_+_34039437 | 0.44 |
ENSMUST00000131134.1
ENSMUST00000087497.4 ENSMUST00000114255.1 ENSMUST00000114252.1 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr9_-_58741543 | 0.44 |
ENSMUST00000098674.4
|
2410076I21Rik
|
RIKEN cDNA 2410076I21 gene |
| chr12_-_110696289 | 0.44 |
ENSMUST00000021698.6
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr1_+_170308802 | 0.43 |
ENSMUST00000056991.5
|
1700015E13Rik
|
RIKEN cDNA 1700015E13 gene |
| chr11_+_74082907 | 0.43 |
ENSMUST00000178159.1
|
Zfp616
|
zinc finger protein 616 |
| chr7_+_17087934 | 0.43 |
ENSMUST00000152671.1
|
Psg16
|
pregnancy specific glycoprotein 16 |
| chr2_+_125068118 | 0.43 |
ENSMUST00000070353.3
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr1_+_40515362 | 0.43 |
ENSMUST00000027237.5
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr4_+_13743424 | 0.43 |
ENSMUST00000006761.3
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr1_-_93342734 | 0.43 |
ENSMUST00000027493.3
|
Pask
|
PAS domain containing serine/threonine kinase |
| chr9_+_119063429 | 0.42 |
ENSMUST00000141185.1
ENSMUST00000126251.1 ENSMUST00000136561.1 |
Vill
|
villin-like |
| chr4_-_140810646 | 0.42 |
ENSMUST00000026377.2
|
Padi3
|
peptidyl arginine deiminase, type III |
| chr2_-_73453918 | 0.42 |
ENSMUST00000102679.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr18_+_69346143 | 0.42 |
ENSMUST00000114980.1
|
Tcf4
|
transcription factor 4 |
| chr8_+_45627946 | 0.42 |
ENSMUST00000145458.1
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr18_+_4920509 | 0.42 |
ENSMUST00000126977.1
|
Svil
|
supervillin |
| chr7_-_104129792 | 0.42 |
ENSMUST00000053743.4
|
4931431F19Rik
|
RIKEN cDNA 4931431F19 gene |
| chr19_-_8819314 | 0.41 |
ENSMUST00000096751.4
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr14_-_47276790 | 0.41 |
ENSMUST00000111792.1
ENSMUST00000111791.1 ENSMUST00000111790.1 |
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr2_+_177508570 | 0.41 |
ENSMUST00000108940.2
|
Gm14403
|
predicted gene 14403 |
| chr13_-_21716143 | 0.41 |
ENSMUST00000091756.1
|
Hist1h2bl
|
histone cluster 1, H2bl |
| chr17_+_47611570 | 0.40 |
ENSMUST00000024778.2
|
Med20
|
mediator complex subunit 20 |
| chr12_+_55199533 | 0.40 |
ENSMUST00000177978.1
|
1700047I17Rik2
|
RIKEN cDNA 1700047I17 gene 2 |
| chr11_+_106216926 | 0.40 |
ENSMUST00000021046.5
|
Ddx42
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 42 |
| chr19_-_8818924 | 0.40 |
ENSMUST00000153281.1
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr18_-_24603464 | 0.39 |
ENSMUST00000154205.1
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr19_-_37176055 | 0.39 |
ENSMUST00000142973.1
ENSMUST00000154376.1 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr4_+_62525369 | 0.39 |
ENSMUST00000062145.1
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr8_-_57653023 | 0.39 |
ENSMUST00000034021.5
|
Galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr16_-_58638739 | 0.39 |
ENSMUST00000053249.8
|
E330017A01Rik
|
RIKEN cDNA E330017A01 gene |
| chrX_-_106221145 | 0.39 |
ENSMUST00000113495.2
|
Taf9b
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of Dlx5_Dlx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.4 | 2.6 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.4 | 1.2 | GO:1904868 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.4 | 2.3 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.4 | 1.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.4 | 1.1 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.3 | 1.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.3 | 1.0 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.3 | 0.9 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.3 | 1.2 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.3 | 0.9 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.3 | 0.6 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.2 | 1.7 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.2 | 0.9 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.2 | 1.1 | GO:0070560 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) protein secretion by platelet(GO:0070560) |
| 0.2 | 0.8 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.2 | 0.8 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.2 | 0.4 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.2 | 0.5 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.2 | 2.4 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.2 | 0.7 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 0.2 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 0.2 | 1.5 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.2 | 0.8 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 0.7 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.2 | 0.6 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 0.8 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 1.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.7 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 1.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.7 | GO:0021828 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 1.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.9 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 | 0.5 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 0.8 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 1.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.4 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.4 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 0.4 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 1.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.5 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 2.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 1.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.4 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 1.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.4 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.7 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.4 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.7 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.6 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.4 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.8 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.4 | GO:0033370 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.1 | 0.3 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.4 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.4 | GO:0070537 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.2 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 1.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 1.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.2 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.1 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.7 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.3 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.2 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.1 | 0.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 2.0 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 0.3 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 1.0 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 0.3 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 2.0 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.1 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 1.3 | GO:0044146 | negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.1 | 0.2 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 | 0.2 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.0 | 1.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.7 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 1.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.5 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 1.3 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 1.7 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.6 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 1.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.4 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0036446 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.3 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.3 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.3 | GO:0002681 | B cell lineage commitment(GO:0002326) somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.1 | GO:0038027 | gonad morphogenesis(GO:0035262) apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.0 | 0.5 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.2 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.1 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.6 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 2.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 5.1 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.5 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.0 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.0 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.3 | GO:0048199 | Golgi to endosome transport(GO:0006895) vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 0.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 1.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.8 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.1 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.6 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.1 | GO:0001805 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.0 | 1.4 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.0 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.1 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.6 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.2 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 2.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 1.8 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1904253 | estrogen biosynthetic process(GO:0006703) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.1 | GO:0035633 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.9 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.0 | 0.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.7 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.5 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.6 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.6 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 1.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.7 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 0.2 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.4 | 1.8 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 0.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 1.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 0.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 1.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.4 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 0.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.2 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 0.8 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.8 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 2.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.3 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 0.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 4.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 2.8 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 1.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.0 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 3.1 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 2.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.0 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.4 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 1.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.9 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 1.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.5 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.8 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 1.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.0 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 0.5 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0004350 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.5 | 1.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 2.6 | GO:0002135 | CTP binding(GO:0002135) |
| 0.4 | 1.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.4 | 1.8 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.4 | 1.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.3 | 2.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 0.9 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.2 | 1.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 0.9 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 2.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 1.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 1.2 | GO:0032407 | MutLalpha complex binding(GO:0032405) MutSalpha complex binding(GO:0032407) |
| 0.2 | 0.6 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.2 | 1.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 0.8 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 0.9 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.6 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 1.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.5 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.3 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 1.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.7 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.2 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.8 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.3 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.2 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.8 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.6 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 1.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.0 | 0.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 5.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 1.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 1.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 1.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 1.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 1.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.0 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.8 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 2.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 0.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 3.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 2.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 4.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.7 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.6 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.5 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 2.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |