Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
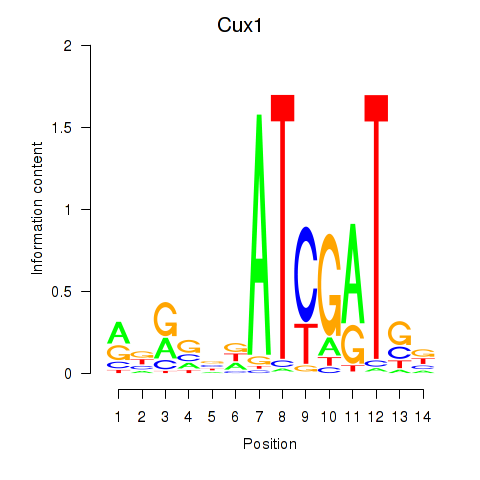
Results for Cux1
Z-value: 1.38
Transcription factors associated with Cux1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cux1
|
ENSMUSG00000029705.11 | cut-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cux1 | mm10_v2_chr5_-_136567307_136567445 | -0.64 | 2.8e-05 | Click! |
Activity profile of Cux1 motif
Sorted Z-values of Cux1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39287074 | 17.89 |
ENSMUST00000003137.8
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr4_+_63344548 | 11.99 |
ENSMUST00000030044.2
|
Orm1
|
orosomucoid 1 |
| chr4_-_62150810 | 10.06 |
ENSMUST00000077719.3
|
Mup21
|
major urinary protein 21 |
| chr13_-_4523322 | 9.53 |
ENSMUST00000080361.5
ENSMUST00000078239.3 |
Akr1c20
|
aldo-keto reductase family 1, member C20 |
| chr15_+_6445320 | 9.42 |
ENSMUST00000022749.9
|
C9
|
complement component 9 |
| chr3_-_81975742 | 8.53 |
ENSMUST00000029645.8
|
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr19_+_39992424 | 8.25 |
ENSMUST00000049178.2
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr7_-_99695809 | 7.92 |
ENSMUST00000107086.2
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr19_+_40089688 | 7.73 |
ENSMUST00000068094.6
ENSMUST00000080171.2 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr7_-_99695628 | 7.29 |
ENSMUST00000145381.1
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr7_-_99695572 | 6.92 |
ENSMUST00000137914.1
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr19_+_39007019 | 6.81 |
ENSMUST00000025966.4
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr19_-_44407703 | 6.58 |
ENSMUST00000041331.2
|
Scd1
|
stearoyl-Coenzyme A desaturase 1 |
| chr7_+_27029074 | 6.56 |
ENSMUST00000075552.5
|
Cyp2a12
|
cytochrome P450, family 2, subfamily a, polypeptide 12 |
| chr7_+_44384098 | 6.49 |
ENSMUST00000118962.1
ENSMUST00000118831.1 |
Syt3
|
synaptotagmin III |
| chr7_+_44384803 | 6.36 |
ENSMUST00000120262.1
|
Syt3
|
synaptotagmin III |
| chr11_+_78499087 | 6.18 |
ENSMUST00000017488.4
|
Vtn
|
vitronectin |
| chr1_+_166254095 | 5.99 |
ENSMUST00000111416.1
|
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr19_-_40073731 | 5.98 |
ENSMUST00000048959.3
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
| chr7_+_27119909 | 5.94 |
ENSMUST00000003100.8
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr2_+_102706356 | 5.84 |
ENSMUST00000123759.1
ENSMUST00000111212.1 ENSMUST00000005220.4 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr13_+_4436094 | 5.51 |
ENSMUST00000156277.1
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr9_+_46268601 | 5.48 |
ENSMUST00000121598.1
|
Apoa5
|
apolipoprotein A-V |
| chr7_-_19698383 | 5.36 |
ENSMUST00000173739.1
|
Apoe
|
apolipoprotein E |
| chr2_-_24049389 | 5.17 |
ENSMUST00000051416.5
|
Hnmt
|
histamine N-methyltransferase |
| chr8_+_109990430 | 4.91 |
ENSMUST00000001720.7
ENSMUST00000143741.1 |
Tat
|
tyrosine aminotransferase |
| chr11_-_72266596 | 4.85 |
ENSMUST00000021161.6
ENSMUST00000140167.1 |
Slc13a5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr7_+_44384604 | 4.83 |
ENSMUST00000130707.1
ENSMUST00000130844.1 |
Syt3
|
synaptotagmin III |
| chr8_-_94696223 | 4.62 |
ENSMUST00000034227.4
|
Pllp
|
plasma membrane proteolipid |
| chr7_-_19698206 | 4.54 |
ENSMUST00000172808.1
ENSMUST00000174191.1 |
Apoe
|
apolipoprotein E |
| chr5_-_89457763 | 4.45 |
ENSMUST00000049209.8
|
Gc
|
group specific component |
| chr10_+_62071014 | 4.41 |
ENSMUST00000053865.5
|
Gm5424
|
predicted gene 5424 |
| chr6_+_121346618 | 4.33 |
ENSMUST00000032200.9
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr9_-_119157055 | 4.25 |
ENSMUST00000010795.4
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr7_+_44590886 | 4.24 |
ENSMUST00000107906.3
|
Kcnc3
|
potassium voltage gated channel, Shaw-related subfamily, member 3 |
| chr11_-_58613481 | 3.97 |
ENSMUST00000048801.7
|
2210407C18Rik
|
RIKEN cDNA 2210407C18 gene |
| chr7_-_140154712 | 3.91 |
ENSMUST00000059241.7
|
Sprn
|
shadow of prion protein |
| chr7_-_14492926 | 3.89 |
ENSMUST00000108524.3
|
Sult2a7
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 7 |
| chr3_+_138374121 | 3.84 |
ENSMUST00000171054.1
|
Adh6-ps1
|
alcohol dehydrogenase 6 (class V), pseudogene 1 |
| chr17_+_24736639 | 3.77 |
ENSMUST00000115262.1
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr12_+_108334341 | 3.73 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr5_-_87254804 | 3.63 |
ENSMUST00000075858.3
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr6_-_128526703 | 3.57 |
ENSMUST00000143664.1
ENSMUST00000112132.1 ENSMUST00000032510.7 |
Pzp
|
pregnancy zone protein |
| chr1_+_58113136 | 3.54 |
ENSMUST00000040999.7
|
Aox3
|
aldehyde oxidase 3 |
| chr19_-_43524462 | 3.53 |
ENSMUST00000026196.7
|
Got1
|
glutamate oxaloacetate transaminase 1, soluble |
| chr7_-_132576372 | 3.52 |
ENSMUST00000084500.6
|
Oat
|
ornithine aminotransferase |
| chr13_-_41847482 | 3.47 |
ENSMUST00000072012.3
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr9_+_114731177 | 3.45 |
ENSMUST00000035007.8
|
Cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr1_-_162898665 | 3.42 |
ENSMUST00000111510.1
ENSMUST00000045902.6 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr1_-_162898484 | 3.39 |
ENSMUST00000143123.1
|
Fmo2
|
flavin containing monooxygenase 2 |
| chr19_-_4839286 | 3.38 |
ENSMUST00000037246.5
|
Ccs
|
copper chaperone for superoxide dismutase |
| chr3_+_94372794 | 3.36 |
ENSMUST00000029795.3
|
Rorc
|
RAR-related orphan receptor gamma |
| chr2_-_166155272 | 3.31 |
ENSMUST00000088086.3
|
Sulf2
|
sulfatase 2 |
| chr6_-_90224438 | 3.29 |
ENSMUST00000076086.2
|
Vmn1r53
|
vomeronasal 1 receptor 53 |
| chr12_-_103904887 | 3.28 |
ENSMUST00000074051.5
|
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1C |
| chr12_-_103773592 | 3.27 |
ENSMUST00000078869.5
|
Serpina1d
|
serine (or cysteine) peptidase inhibitor, clade A, member 1D |
| chr11_-_50325599 | 3.17 |
ENSMUST00000179865.1
ENSMUST00000020637.8 |
Canx
|
calnexin |
| chrM_+_10167 | 3.16 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr7_-_26939377 | 3.14 |
ENSMUST00000170227.1
|
Cyp2a22
|
cytochrome P450, family 2, subfamily a, polypeptide 22 |
| chr1_+_78511865 | 3.13 |
ENSMUST00000012331.6
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr18_-_38211957 | 3.12 |
ENSMUST00000159405.1
ENSMUST00000160721.1 |
Pcdh1
|
protocadherin 1 |
| chr7_-_35215248 | 3.07 |
ENSMUST00000118444.1
ENSMUST00000122409.1 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr2_+_31887262 | 3.06 |
ENSMUST00000138325.1
ENSMUST00000028187.6 |
Lamc3
|
laminin gamma 3 |
| chr17_+_24736673 | 3.03 |
ENSMUST00000101800.5
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr13_+_4059565 | 2.98 |
ENSMUST00000041768.6
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr5_-_87140318 | 2.96 |
ENSMUST00000067790.6
ENSMUST00000113327.1 |
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr11_-_116198701 | 2.95 |
ENSMUST00000072948.4
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr14_-_47189406 | 2.90 |
ENSMUST00000089959.6
|
Gch1
|
GTP cyclohydrolase 1 |
| chr2_-_164638789 | 2.87 |
ENSMUST00000109336.1
|
Wfdc16
|
WAP four-disulfide core domain 16 |
| chr11_+_75468040 | 2.85 |
ENSMUST00000043598.7
ENSMUST00000108435.1 |
Tlcd2
|
TLC domain containing 2 |
| chr11_-_96916366 | 2.84 |
ENSMUST00000144731.1
ENSMUST00000127048.1 |
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr2_-_86347764 | 2.80 |
ENSMUST00000099894.2
|
Olfr1055
|
olfactory receptor 1055 |
| chrX_+_101383726 | 2.80 |
ENSMUST00000119190.1
|
Gjb1
|
gap junction protein, beta 1 |
| chr2_-_32694120 | 2.75 |
ENSMUST00000028148.4
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr13_+_93674403 | 2.72 |
ENSMUST00000048001.6
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr5_-_147894804 | 2.68 |
ENSMUST00000118527.1
ENSMUST00000031655.3 ENSMUST00000138244.1 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr11_-_96916448 | 2.68 |
ENSMUST00000103152.4
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr11_-_74925925 | 2.65 |
ENSMUST00000121738.1
|
Srr
|
serine racemase |
| chrX_-_8193387 | 2.65 |
ENSMUST00000143223.1
ENSMUST00000033509.8 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr12_-_103863551 | 2.63 |
ENSMUST00000085056.6
ENSMUST00000072876.5 ENSMUST00000124717.1 |
Serpina1a
|
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
| chr5_+_33104219 | 2.63 |
ENSMUST00000011178.2
|
Slc5a1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr10_-_75780954 | 2.62 |
ENSMUST00000173512.1
ENSMUST00000173537.1 |
Gm20441
Gstt3
|
predicted gene 20441 glutathione S-transferase, theta 3 |
| chr19_+_45076105 | 2.60 |
ENSMUST00000026234.4
|
Kazald1
|
Kazal-type serine peptidase inhibitor domain 1 |
| chr4_-_63154130 | 2.58 |
ENSMUST00000030041.4
|
Ambp
|
alpha 1 microglobulin/bikunin |
| chrX_+_101377267 | 2.54 |
ENSMUST00000052130.7
|
Gjb1
|
gap junction protein, beta 1 |
| chr11_+_51763682 | 2.54 |
ENSMUST00000020653.5
|
Sar1b
|
SAR1 gene homolog B (S. cerevisiae) |
| chr11_-_50931612 | 2.53 |
ENSMUST00000109124.3
|
Zfp354b
|
zinc finger protein 354B |
| chr4_+_62360695 | 2.50 |
ENSMUST00000084526.5
|
Slc31a1
|
solute carrier family 31, member 1 |
| chr13_-_34130345 | 2.49 |
ENSMUST00000075774.3
|
Tubb2b
|
tubulin, beta 2B class IIB |
| chr1_+_88070765 | 2.47 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr1_-_180256294 | 2.44 |
ENSMUST00000111108.3
|
Psen2
|
presenilin 2 |
| chrX_+_142228177 | 2.42 |
ENSMUST00000112914.1
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr15_+_88819584 | 2.34 |
ENSMUST00000024042.3
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr1_+_88200601 | 2.33 |
ENSMUST00000049289.8
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr5_+_3596066 | 2.33 |
ENSMUST00000006061.6
ENSMUST00000121291.1 ENSMUST00000142516.1 |
Pex1
|
peroxisomal biogenesis factor 1 |
| chr12_-_103738158 | 2.32 |
ENSMUST00000095450.4
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr11_-_96916407 | 2.30 |
ENSMUST00000130774.1
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr8_-_77610597 | 2.29 |
ENSMUST00000034030.8
|
Tmem184c
|
transmembrane protein 184C |
| chr7_-_27674516 | 2.28 |
ENSMUST00000036453.7
ENSMUST00000108341.1 |
Map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr12_+_24572276 | 2.27 |
ENSMUST00000085553.5
|
Grhl1
|
grainyhead-like 1 (Drosophila) |
| chr9_-_50746501 | 2.26 |
ENSMUST00000034564.1
|
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr1_-_179546261 | 2.23 |
ENSMUST00000027769.5
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr9_-_26999491 | 2.22 |
ENSMUST00000060513.7
ENSMUST00000120367.1 |
Acad8
|
acyl-Coenzyme A dehydrogenase family, member 8 |
| chr4_+_104766334 | 2.21 |
ENSMUST00000065072.6
|
C8b
|
complement component 8, beta polypeptide |
| chr6_+_88724828 | 2.19 |
ENSMUST00000089449.2
|
Mgll
|
monoglyceride lipase |
| chr13_-_34963788 | 2.18 |
ENSMUST00000164155.1
ENSMUST00000021853.5 |
Eci3
|
enoyl-Coenzyme A delta isomerase 3 |
| chr19_-_39649046 | 2.18 |
ENSMUST00000067328.6
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chrX_+_7722267 | 2.17 |
ENSMUST00000125991.1
ENSMUST00000148624.1 |
Wdr45
|
WD repeat domain 45 |
| chr6_-_124741374 | 2.16 |
ENSMUST00000004389.5
|
Grcc10
|
gene rich cluster, C10 gene |
| chr5_-_87424201 | 2.16 |
ENSMUST00000072818.5
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr4_-_148160031 | 2.16 |
ENSMUST00000057907.3
|
Fbxo44
|
F-box protein 44 |
| chr1_+_16688405 | 2.16 |
ENSMUST00000026881.4
|
Ly96
|
lymphocyte antigen 96 |
| chr11_+_73199445 | 2.15 |
ENSMUST00000006105.6
|
Shpk
|
sedoheptulokinase |
| chr6_-_114969986 | 2.15 |
ENSMUST00000139640.1
|
Vgll4
|
vestigial like 4 (Drosophila) |
| chr6_+_48395586 | 2.15 |
ENSMUST00000114571.1
ENSMUST00000114572.3 ENSMUST00000031815.5 |
Krba1
|
KRAB-A domain containing 1 |
| chrX_+_142227923 | 2.12 |
ENSMUST00000042329.5
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr11_-_53423123 | 2.12 |
ENSMUST00000036045.5
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr2_-_84775388 | 2.10 |
ENSMUST00000023994.3
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr15_+_99392948 | 2.09 |
ENSMUST00000161250.1
ENSMUST00000160635.1 ENSMUST00000161778.1 |
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr10_-_127121125 | 2.09 |
ENSMUST00000164259.1
ENSMUST00000080975.4 |
Os9
|
amplified in osteosarcoma |
| chrM_+_7005 | 2.08 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr2_-_84775420 | 2.08 |
ENSMUST00000111641.1
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chrX_+_7722214 | 2.07 |
ENSMUST00000043045.2
ENSMUST00000116634.1 ENSMUST00000115689.3 ENSMUST00000131077.1 ENSMUST00000115688.1 ENSMUST00000116633.1 |
Wdr45
|
WD repeat domain 45 |
| chr8_+_94525067 | 1.98 |
ENSMUST00000098489.4
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr7_+_24587543 | 1.97 |
ENSMUST00000077191.6
|
Ethe1
|
ethylmalonic encephalopathy 1 |
| chr3_-_85722474 | 1.94 |
ENSMUST00000119077.1
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr9_-_101034857 | 1.93 |
ENSMUST00000142676.1
ENSMUST00000149322.1 |
Pccb
|
propionyl Coenzyme A carboxylase, beta polypeptide |
| chr3_-_107239707 | 1.92 |
ENSMUST00000049852.8
|
Prok1
|
prokineticin 1 |
| chr14_+_118137101 | 1.91 |
ENSMUST00000022728.2
|
Gpr180
|
G protein-coupled receptor 180 |
| chr6_+_48395652 | 1.90 |
ENSMUST00000077093.4
|
Krba1
|
KRAB-A domain containing 1 |
| chr3_+_19957037 | 1.89 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chrM_+_9870 | 1.86 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr8_-_77610668 | 1.85 |
ENSMUST00000141202.1
ENSMUST00000152168.1 |
Tmem184c
|
transmembrane protein 184C |
| chr9_-_101034892 | 1.85 |
ENSMUST00000035116.5
|
Pccb
|
propionyl Coenzyme A carboxylase, beta polypeptide |
| chr11_-_106487796 | 1.78 |
ENSMUST00000001059.2
ENSMUST00000106799.1 ENSMUST00000106800.1 |
Ern1
|
endoplasmic reticulum (ER) to nucleus signalling 1 |
| chr6_+_124931378 | 1.78 |
ENSMUST00000032214.7
ENSMUST00000180095.1 |
Mlf2
|
myeloid leukemia factor 2 |
| chr5_+_90561102 | 1.77 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr4_+_43493345 | 1.76 |
ENSMUST00000030181.5
ENSMUST00000107922.2 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr4_+_104766308 | 1.75 |
ENSMUST00000031663.3
|
C8b
|
complement component 8, beta polypeptide |
| chr11_-_101171302 | 1.74 |
ENSMUST00000164474.1
ENSMUST00000043397.7 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr3_-_89279633 | 1.74 |
ENSMUST00000118860.1
ENSMUST00000029566.2 |
Efna1
|
ephrin A1 |
| chr12_+_55598917 | 1.73 |
ENSMUST00000051857.3
|
Insm2
|
insulinoma-associated 2 |
| chr9_+_46240696 | 1.72 |
ENSMUST00000034585.6
|
Apoa4
|
apolipoprotein A-IV |
| chr1_+_21218575 | 1.71 |
ENSMUST00000027065.5
ENSMUST00000027064.7 |
Tmem14a
|
transmembrane protein 14A |
| chr1_-_84284548 | 1.71 |
ENSMUST00000177458.1
ENSMUST00000168574.2 |
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr17_-_23786046 | 1.70 |
ENSMUST00000024704.3
|
Flywch2
|
FLYWCH family member 2 |
| chr4_-_141933080 | 1.70 |
ENSMUST00000036701.7
|
Fhad1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr18_-_56562261 | 1.70 |
ENSMUST00000066208.6
ENSMUST00000172734.1 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr19_-_42128982 | 1.69 |
ENSMUST00000161873.1
|
Avpi1
|
arginine vasopressin-induced 1 |
| chr1_-_24612700 | 1.69 |
ENSMUST00000088336.1
|
Gm10222
|
predicted gene 10222 |
| chrX_+_36795642 | 1.69 |
ENSMUST00000016463.3
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr4_+_116596672 | 1.68 |
ENSMUST00000051869.7
|
Ccdc17
|
coiled-coil domain containing 17 |
| chr1_+_78511805 | 1.68 |
ENSMUST00000152111.1
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr6_-_130231638 | 1.68 |
ENSMUST00000088011.4
ENSMUST00000112013.1 ENSMUST00000049304.7 |
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7 |
| chr3_-_88296838 | 1.67 |
ENSMUST00000010682.3
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr3_-_63964659 | 1.67 |
ENSMUST00000161659.1
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr17_+_46711459 | 1.66 |
ENSMUST00000002840.8
|
Pex6
|
peroxisomal biogenesis factor 6 |
| chr3_+_19957088 | 1.66 |
ENSMUST00000108328.1
|
Cp
|
ceruloplasmin |
| chr7_-_137410717 | 1.65 |
ENSMUST00000120340.1
ENSMUST00000117404.1 ENSMUST00000068996.6 |
9430038I01Rik
|
RIKEN cDNA 9430038I01 gene |
| chr1_+_106171752 | 1.64 |
ENSMUST00000061047.6
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr3_-_88296979 | 1.61 |
ENSMUST00000107556.3
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr5_+_24428208 | 1.59 |
ENSMUST00000115049.2
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr17_-_24220738 | 1.58 |
ENSMUST00000024930.7
|
1600002H07Rik
|
RIKEN cDNA 1600002H07 gene |
| chr7_-_101921186 | 1.56 |
ENSMUST00000106965.1
ENSMUST00000106968.1 ENSMUST00000106967.1 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr11_+_96251100 | 1.55 |
ENSMUST00000129907.2
|
Gm53
|
predicted gene 53 |
| chr15_+_76660564 | 1.54 |
ENSMUST00000004294.10
|
Kifc2
|
kinesin family member C2 |
| chr3_+_19957240 | 1.54 |
ENSMUST00000108325.2
|
Cp
|
ceruloplasmin |
| chrX_-_60893430 | 1.52 |
ENSMUST00000135107.2
|
Sox3
|
SRY-box containing gene 3 |
| chr4_-_148151646 | 1.51 |
ENSMUST00000132083.1
|
Fbxo6
|
F-box protein 6 |
| chr14_-_59365465 | 1.50 |
ENSMUST00000095157.4
|
Phf11d
|
PHD finger protein 11D |
| chrX_+_152001845 | 1.50 |
ENSMUST00000026289.3
ENSMUST00000112617.3 |
Hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr5_+_124541296 | 1.49 |
ENSMUST00000124529.1
|
Tmed2
|
transmembrane emp24 domain trafficking protein 2 |
| chr18_+_37447641 | 1.48 |
ENSMUST00000052387.3
|
Pcdhb14
|
protocadherin beta 14 |
| chr2_-_110305730 | 1.47 |
ENSMUST00000046233.2
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr11_-_105944128 | 1.46 |
ENSMUST00000184086.1
|
Cyb561
|
cytochrome b-561 |
| chr16_+_70314087 | 1.44 |
ENSMUST00000023393.8
|
Gbe1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr12_+_57564111 | 1.42 |
ENSMUST00000101398.3
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr7_-_24587612 | 1.42 |
ENSMUST00000094705.2
|
Zfp575
|
zinc finger protein 575 |
| chr12_-_103956891 | 1.42 |
ENSMUST00000085054.4
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr19_+_43782181 | 1.41 |
ENSMUST00000026208.4
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr8_+_11497506 | 1.41 |
ENSMUST00000177955.1
ENSMUST00000033901.4 ENSMUST00000178721.1 |
Carkd
|
carbohydrate kinase domain containing |
| chr7_-_140881811 | 1.40 |
ENSMUST00000106048.3
ENSMUST00000147331.2 ENSMUST00000137710.1 |
Sirt3
|
sirtuin 3 |
| chr1_+_107361929 | 1.40 |
ENSMUST00000027566.2
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr4_-_118134869 | 1.40 |
ENSMUST00000097912.1
ENSMUST00000030263.2 ENSMUST00000106410.1 |
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr6_-_24527546 | 1.39 |
ENSMUST00000118558.1
|
Ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr10_+_125966214 | 1.39 |
ENSMUST00000074807.6
|
Lrig3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr6_+_48554776 | 1.39 |
ENSMUST00000114545.1
ENSMUST00000153222.1 ENSMUST00000101436.2 |
Lrrc61
|
leucine rich repeat containing 61 |
| chr10_-_42478488 | 1.38 |
ENSMUST00000041024.8
|
Lace1
|
lactation elevated 1 |
| chr13_+_55714624 | 1.38 |
ENSMUST00000021959.9
|
Txndc15
|
thioredoxin domain containing 15 |
| chr11_-_96075581 | 1.38 |
ENSMUST00000107686.1
ENSMUST00000107684.1 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c1 (subunit 9) |
| chrM_+_7759 | 1.37 |
ENSMUST00000082407.1
ENSMUST00000082408.1 |
mt-Atp8
mt-Atp6
|
mitochondrially encoded ATP synthase 8 mitochondrially encoded ATP synthase 6 |
| chr9_-_50727921 | 1.37 |
ENSMUST00000118707.1
ENSMUST00000034566.8 |
Dixdc1
|
DIX domain containing 1 |
| chr7_+_67647405 | 1.37 |
ENSMUST00000032774.8
ENSMUST00000107471.1 |
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr4_+_95557494 | 1.36 |
ENSMUST00000079223.4
ENSMUST00000177394.1 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr16_+_29579331 | 1.36 |
ENSMUST00000160597.1
|
Opa1
|
optic atrophy 1 |
| chr9_+_107578887 | 1.34 |
ENSMUST00000149638.1
ENSMUST00000139274.1 ENSMUST00000130053.1 ENSMUST00000122985.1 ENSMUST00000127380.1 ENSMUST00000139581.1 |
Nat6
|
N-acetyltransferase 6 |
| chr11_+_108921648 | 1.34 |
ENSMUST00000144511.1
|
Axin2
|
axin2 |
| chr7_+_113207465 | 1.33 |
ENSMUST00000047321.7
|
Arntl
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr15_+_44196135 | 1.33 |
ENSMUST00000038856.6
ENSMUST00000110289.3 |
Trhr
|
thyrotropin releasing hormone receptor |
| chr4_-_91399984 | 1.32 |
ENSMUST00000102799.3
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cux1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 22.1 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 3.3 | 9.9 | GO:1903002 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 2.8 | 8.5 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 1.9 | 62.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.8 | 5.5 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.7 | 6.8 | GO:0030091 | protein repair(GO:0030091) |
| 1.6 | 6.6 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.4 | 4.2 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.4 | 5.5 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 1.2 | 4.9 | GO:0015744 | succinate transport(GO:0015744) |
| 1.2 | 4.8 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.2 | 3.5 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 1.1 | 4.3 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 1.1 | 6.4 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 1.0 | 5.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 1.0 | 9.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 1.0 | 4.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.9 | 2.8 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.9 | 9.5 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.8 | 13.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.8 | 5.8 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.7 | 5.2 | GO:0030969 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.7 | 2.9 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.7 | 15.0 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.7 | 3.5 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.6 | 2.5 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.6 | 4.6 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.6 | 1.7 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) multicellular organism lipid catabolic process(GO:0044240) |
| 0.6 | 1.7 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.5 | 2.2 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.5 | 2.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.5 | 2.2 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.5 | 2.7 | GO:0046416 | D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.5 | 3.5 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.5 | 2.9 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.5 | 5.3 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.5 | 1.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.4 | 1.3 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.4 | 2.7 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.4 | 1.7 | GO:0014028 | notochord formation(GO:0014028) |
| 0.4 | 1.3 | GO:0035602 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) |
| 0.4 | 1.3 | GO:1902871 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.4 | 4.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.4 | 3.7 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.4 | 1.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.4 | 1.4 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.3 | 1.7 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.3 | 2.7 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.3 | 1.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 0.9 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.3 | 2.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 1.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.3 | 3.4 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.3 | 1.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 | 2.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 0.9 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.3 | 16.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.3 | 0.9 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.3 | 0.9 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.3 | 4.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.3 | 4.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.3 | 0.8 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.3 | 1.3 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.3 | 1.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.3 | 1.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.3 | 2.6 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.3 | 0.8 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 2.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 2.0 | GO:0045345 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.2 | 2.0 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.2 | 2.2 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.2 | 1.7 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.2 | 0.7 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.2 | 0.9 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.2 | 2.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 0.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 2.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 2.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.2 | 0.9 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 0.6 | GO:0016131 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.2 | 0.6 | GO:1905035 | regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.2 | 1.0 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.2 | 1.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 0.6 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 3.8 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.2 | 0.8 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.2 | 5.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.2 | 1.1 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.2 | 0.9 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 0.7 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 0.7 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.2 | 1.5 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 2.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.2 | 5.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 1.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 2.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.2 | 0.9 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 2.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 1.0 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 0.7 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 0.7 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 0.3 | GO:0060460 | left lung development(GO:0060459) left lung morphogenesis(GO:0060460) |
| 0.1 | 2.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.8 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.4 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 2.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 9.3 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 0.5 | GO:1903542 | protein to membrane docking(GO:0022615) negative regulation of exosomal secretion(GO:1903542) |
| 0.1 | 0.4 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 1.0 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.4 | GO:0099548 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 1.0 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 3.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 4.4 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 1.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.7 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 1.0 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.4 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.1 | 0.9 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.1 | 0.6 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.5 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 0.3 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.1 | 2.4 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 1.5 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.3 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.9 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 3.4 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.1 | 0.2 | GO:0042697 | menopause(GO:0042697) |
| 0.1 | 1.5 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.2 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.6 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.6 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 7.0 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.1 | 0.6 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.4 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 5.7 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.3 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.1 | 0.2 | GO:0001806 | type IV hypersensitivity(GO:0001806) |
| 0.1 | 0.8 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.3 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.1 | 1.0 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.7 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 3.2 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 1.2 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.4 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.1 | 0.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 1.0 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.1 | 0.5 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.6 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.1 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.3 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.1 | 2.6 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 0.3 | GO:0033505 | ventral midline development(GO:0007418) floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 0.2 | GO:0032430 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.1 | 2.9 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 3.4 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.3 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.5 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.6 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.3 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 4.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.6 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.0 | 4.7 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.9 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 3.3 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.2 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 2.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.7 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.6 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 7.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 2.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.8 | GO:0090662 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.4 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.4 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.6 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.0 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.8 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.9 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.4 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 0.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.2 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.5 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.2 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.5 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.4 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.4 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 2.0 | 9.9 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.9 | 6.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.5 | 2.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.4 | 2.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.4 | 3.1 | GO:1990604 | AIP1-IRE1 complex(GO:1990597) IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.4 | 1.3 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.4 | 1.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.4 | 4.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 4.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.4 | 6.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.4 | 5.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 1.7 | GO:0030891 | VCB complex(GO:0030891) |
| 0.3 | 3.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.3 | 1.7 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.3 | 2.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 23.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 1.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 10.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 5.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 1.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 2.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 4.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 6.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.3 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 10.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 1.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 1.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.1 | 2.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.5 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 6.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 7.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 3.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.0 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.3 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.1 | 0.7 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 0.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 8.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 89.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 3.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.8 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 4.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 5.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 3.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 3.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.5 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 3.3 | 9.9 | GO:0046911 | metal chelating activity(GO:0046911) |
| 3.1 | 18.6 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 2.8 | 8.4 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 2.1 | 8.5 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.8 | 5.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.6 | 6.6 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 1.6 | 4.9 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 1.5 | 4.4 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 1.4 | 7.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 1.3 | 3.8 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 1.2 | 4.8 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 1.1 | 6.8 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 1.1 | 23.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 1.0 | 3.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 1.0 | 5.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.9 | 2.7 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.9 | 23.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.7 | 4.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.7 | 3.5 | GO:0004854 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.7 | 6.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.7 | 2.7 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.7 | 3.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.7 | 3.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.7 | 3.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.6 | 3.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.6 | 2.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.6 | 2.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.5 | 5.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 2.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.5 | 3.0 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.5 | 2.9 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.5 | 1.5 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.5 | 2.9 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.5 | 1.4 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.5 | 5.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 2.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.4 | 1.7 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.4 | 9.1 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.4 | 3.6 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.4 | 3.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.4 | 1.4 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.4 | 13.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 2.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.3 | 1.0 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.3 | 5.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 2.6 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.3 | 4.2 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.3 | 7.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.3 | 2.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 1.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.3 | 0.9 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.3 | 0.9 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.3 | 8.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 15.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.3 | 0.8 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.3 | 3.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 1.5 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.2 | 0.7 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.2 | 3.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 6.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 0.6 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.2 | 5.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 0.6 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.2 | 1.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 0.7 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 2.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 0.9 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 2.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 2.6 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.2 | 0.9 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 0.9 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.2 | 0.6 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.2 | 1.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 3.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.0 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 3.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 4.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.0 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 3.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 1.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 2.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 15.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 2.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.3 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 2.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 2.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 1.8 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.3 | GO:0038100 | nodal binding(GO:0038100) |
| 0.1 | 2.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.3 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 1.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.2 | GO:0001566 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.1 | 0.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.3 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.4 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 1.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 2.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 2.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.4 | GO:0033265 | choline transmembrane transporter activity(GO:0015220) choline binding(GO:0033265) |
| 0.1 | 1.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 2.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 1.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 2.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.3 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 3.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 2.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 1.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.0 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.3 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 3.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 2.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.6 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 3.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 4.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 3.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 14.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 22.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.7 | 17.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.5 | 8.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 14.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 4.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 3.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 3.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 5.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 3.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 5.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 3.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 4.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 3.4 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.2 | 3.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 7.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 2.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 4.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 3.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 4.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 3.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 2.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 3.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 3.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.7 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 8.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 1.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 2.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.7 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 3.3 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 1.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 2.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 2.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 5.6 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 2.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.9 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 0.2 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |