Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Crx_Gsc
Z-value: 0.81
Transcription factors associated with Crx_Gsc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
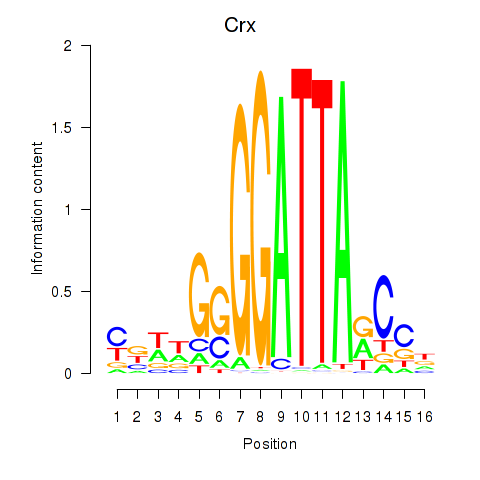
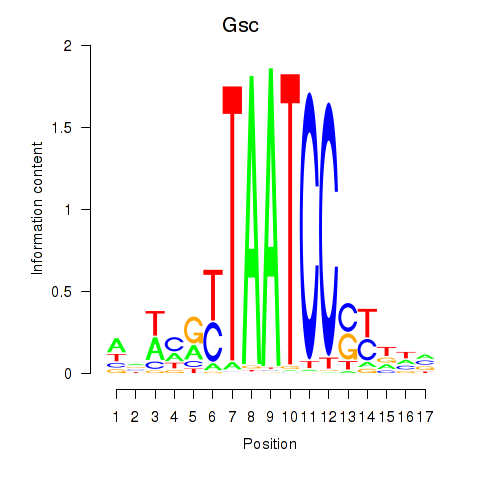
Crx
|
ENSMUSG00000041578.9 | cone-rod homeobox |
|
Gsc
|
ENSMUSG00000021095.4 | goosecoid homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gsc | mm10_v2_chr12_-_104473236_104473324 | 0.34 | 4.0e-02 | Click! |
| Crx | mm10_v2_chr7_-_15879844_15879968 | 0.04 | 8.3e-01 | Click! |
Activity profile of Crx_Gsc motif
Sorted Z-values of Crx_Gsc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_87141114 | 4.82 |
ENSMUST00000168889.1
|
Diap3
|
diaphanous homolog 3 (Drosophila) |
| chr7_-_103843154 | 4.45 |
ENSMUST00000063957.4
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr14_-_87141206 | 4.17 |
ENSMUST00000022599.7
|
Diap3
|
diaphanous homolog 3 (Drosophila) |
| chr14_-_20269162 | 4.08 |
ENSMUST00000024155.7
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr4_-_137430517 | 3.82 |
ENSMUST00000102522.4
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr5_+_90768511 | 3.40 |
ENSMUST00000031319.6
|
Ppbp
|
pro-platelet basic protein |
| chr6_+_113531675 | 3.33 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr3_-_113291449 | 3.29 |
ENSMUST00000179568.1
|
Amy2a4
|
amylase 2a4 |
| chr3_-_113324052 | 3.15 |
ENSMUST00000179314.1
|
Amy2a3
|
amylase 2a3 |
| chr13_-_4150628 | 3.10 |
ENSMUST00000110704.2
ENSMUST00000021635.7 |
Akr1c18
|
aldo-keto reductase family 1, member C18 |
| chr11_-_102107822 | 2.92 |
ENSMUST00000177304.1
ENSMUST00000017455.8 |
Pyy
|
peptide YY |
| chr4_-_137409777 | 2.63 |
ENSMUST00000024200.6
|
Gm13011
|
predicted gene 13011 |
| chr3_-_113258837 | 2.59 |
ENSMUST00000098673.3
|
Amy2a5
|
amylase 2a5 |
| chr14_-_19977151 | 2.58 |
ENSMUST00000055100.7
ENSMUST00000162425.1 |
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr6_-_41035501 | 2.55 |
ENSMUST00000031931.5
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr16_+_33794008 | 2.53 |
ENSMUST00000115044.1
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr14_-_19977040 | 2.50 |
ENSMUST00000159028.1
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr16_+_33794345 | 2.47 |
ENSMUST00000023520.6
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr8_+_72761868 | 2.39 |
ENSMUST00000058099.8
|
F2rl3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr5_+_76656512 | 2.31 |
ENSMUST00000086909.4
|
Gm10430
|
predicted gene 10430 |
| chr14_+_27000362 | 2.27 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr3_+_105870858 | 2.02 |
ENSMUST00000164730.1
|
Adora3
|
adenosine A3 receptor |
| chr4_-_63172118 | 1.99 |
ENSMUST00000030042.2
|
Kif12
|
kinesin family member 12 |
| chr8_+_117498272 | 1.98 |
ENSMUST00000081232.7
|
Plcg2
|
phospholipase C, gamma 2 |
| chr11_-_17008647 | 1.93 |
ENSMUST00000102881.3
|
Plek
|
pleckstrin |
| chr4_-_34050038 | 1.88 |
ENSMUST00000084734.4
|
Spaca1
|
sperm acrosome associated 1 |
| chr2_-_24763047 | 1.83 |
ENSMUST00000100348.3
ENSMUST00000041342.5 ENSMUST00000114447.1 ENSMUST00000102939.2 ENSMUST00000070864.7 |
Cacna1b
|
calcium channel, voltage-dependent, N type, alpha 1B subunit |
| chr18_-_74207771 | 1.76 |
ENSMUST00000040188.8
ENSMUST00000177604.1 |
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr14_-_19977249 | 1.71 |
ENSMUST00000160013.1
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr4_-_141078302 | 1.66 |
ENSMUST00000030760.8
|
Necap2
|
NECAP endocytosis associated 2 |
| chr11_+_95010277 | 1.63 |
ENSMUST00000124735.1
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr4_-_34050077 | 1.60 |
ENSMUST00000029927.5
|
Spaca1
|
sperm acrosome associated 1 |
| chr7_-_110982169 | 1.53 |
ENSMUST00000154466.1
|
Mrvi1
|
MRV integration site 1 |
| chr6_-_136401830 | 1.49 |
ENSMUST00000058713.7
|
E330021D16Rik
|
RIKEN cDNA E330021D16 gene |
| chr3_+_105870898 | 1.49 |
ENSMUST00000010279.5
|
Adora3
|
adenosine A3 receptor |
| chr2_+_178430531 | 1.48 |
ENSMUST00000108912.2
ENSMUST00000042092.8 |
Cdh26
|
cadherin-like 26 |
| chr14_+_65266701 | 1.46 |
ENSMUST00000169656.1
|
Fbxo16
|
F-box protein 16 |
| chr1_-_189688074 | 1.45 |
ENSMUST00000171929.1
ENSMUST00000165962.1 |
Cenpf
|
centromere protein F |
| chr7_+_99535652 | 1.32 |
ENSMUST00000032995.8
ENSMUST00000162404.1 |
Arrb1
|
arrestin, beta 1 |
| chrX_+_7919816 | 1.32 |
ENSMUST00000041096.3
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr7_+_99535439 | 1.32 |
ENSMUST00000098266.2
ENSMUST00000179755.1 |
Arrb1
|
arrestin, beta 1 |
| chr8_+_123411424 | 1.31 |
ENSMUST00000071134.3
|
Tubb3
|
tubulin, beta 3 class III |
| chr7_-_43533171 | 1.26 |
ENSMUST00000004728.5
ENSMUST00000039861.5 |
Cd33
|
CD33 antigen |
| chr2_+_120476911 | 1.22 |
ENSMUST00000110716.1
ENSMUST00000028748.6 ENSMUST00000090028.5 ENSMUST00000110719.2 |
Capn3
|
calpain 3 |
| chr6_-_128826305 | 1.22 |
ENSMUST00000174544.1
ENSMUST00000172887.1 ENSMUST00000032472.4 |
Klrb1b
|
killer cell lectin-like receptor subfamily B member 1B |
| chr2_-_160912292 | 1.21 |
ENSMUST00000109454.1
ENSMUST00000057169.4 |
Emilin3
|
elastin microfibril interfacer 3 |
| chr7_-_4752972 | 1.18 |
ENSMUST00000183971.1
ENSMUST00000182173.1 ENSMUST00000182738.1 ENSMUST00000184143.1 ENSMUST00000182111.1 ENSMUST00000182048.1 ENSMUST00000063324.7 |
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr9_-_53975246 | 1.12 |
ENSMUST00000048409.7
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr10_+_3973086 | 1.11 |
ENSMUST00000117291.1
ENSMUST00000120585.1 ENSMUST00000043735.7 |
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr2_-_164404606 | 1.10 |
ENSMUST00000109359.1
ENSMUST00000109358.1 ENSMUST00000103103.3 |
Matn4
|
matrilin 4 |
| chr10_-_62327757 | 1.09 |
ENSMUST00000139228.1
|
Hk1
|
hexokinase 1 |
| chr16_-_76022266 | 1.02 |
ENSMUST00000114240.1
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr11_+_67277124 | 1.01 |
ENSMUST00000019625.5
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr7_-_45092198 | 1.01 |
ENSMUST00000140449.1
ENSMUST00000117546.1 ENSMUST00000019683.3 |
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr5_-_38876693 | 0.97 |
ENSMUST00000169819.1
ENSMUST00000171633.1 |
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr9_-_95750335 | 0.96 |
ENSMUST00000053785.3
|
Trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr15_+_85017138 | 0.96 |
ENSMUST00000023070.5
|
Upk3a
|
uroplakin 3A |
| chr5_+_13399309 | 0.96 |
ENSMUST00000030714.7
ENSMUST00000141968.1 |
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr8_-_94876269 | 0.90 |
ENSMUST00000046461.7
|
Dok4
|
docking protein 4 |
| chr19_+_3323301 | 0.87 |
ENSMUST00000025835.4
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr9_+_27030159 | 0.87 |
ENSMUST00000073127.7
ENSMUST00000086198.4 |
Ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr12_-_118198917 | 0.86 |
ENSMUST00000084806.6
|
Dnah11
|
dynein, axonemal, heavy chain 11 |
| chr9_+_108991902 | 0.85 |
ENSMUST00000147989.1
ENSMUST00000051873.8 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr12_-_115964196 | 0.85 |
ENSMUST00000103550.2
|
Ighv1-83
|
immunoglobulin heavy variable 1-83 |
| chr8_+_45885479 | 0.85 |
ENSMUST00000034053.5
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr16_+_32186192 | 0.84 |
ENSMUST00000099990.3
|
Bex6
|
brain expressed gene 6 |
| chr1_+_74791516 | 0.83 |
ENSMUST00000006718.8
|
Wnt10a
|
wingless related MMTV integration site 10a |
| chr11_-_120598346 | 0.83 |
ENSMUST00000026125.2
|
Alyref
|
Aly/REF export factor |
| chr17_+_37002510 | 0.83 |
ENSMUST00000069250.7
|
Zfp57
|
zinc finger protein 57 |
| chr2_-_101883010 | 0.83 |
ENSMUST00000154525.1
|
Prr5l
|
proline rich 5 like |
| chr18_-_70141568 | 0.82 |
ENSMUST00000121693.1
|
Rab27b
|
RAB27b, member RAS oncogene family |
| chr3_+_28781305 | 0.82 |
ENSMUST00000060500.7
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr10_-_62379852 | 0.81 |
ENSMUST00000143236.1
ENSMUST00000133429.1 ENSMUST00000132926.1 ENSMUST00000116238.2 |
Hk1
|
hexokinase 1 |
| chr12_+_55124528 | 0.80 |
ENSMUST00000177768.1
|
Fam177a
|
family with sequence similarity 177, member A |
| chr7_-_45092130 | 0.80 |
ENSMUST00000148175.1
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr6_-_136922169 | 0.79 |
ENSMUST00000032343.6
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr1_+_178187721 | 0.75 |
ENSMUST00000159284.1
|
Desi2
|
desumoylating isopeptidase 2 |
| chrX_+_164140447 | 0.74 |
ENSMUST00000073973.4
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr13_+_21811737 | 0.73 |
ENSMUST00000104941.2
|
Hist1h4m
|
histone cluster 1, H4m |
| chr5_-_5694559 | 0.72 |
ENSMUST00000115426.2
|
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr17_+_35821675 | 0.72 |
ENSMUST00000003635.6
|
Ier3
|
immediate early response 3 |
| chr1_-_149922339 | 0.72 |
ENSMUST00000111926.2
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr8_-_105707933 | 0.71 |
ENSMUST00000013299.9
|
Enkd1
|
enkurin domain containing 1 |
| chr3_-_59344256 | 0.71 |
ENSMUST00000039419.6
|
Igsf10
|
immunoglobulin superfamily, member 10 |
| chr7_+_30787897 | 0.71 |
ENSMUST00000098559.1
|
Krtdap
|
keratinocyte differentiation associated protein |
| chr19_-_7711263 | 0.69 |
ENSMUST00000025666.7
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chrX_+_153139941 | 0.68 |
ENSMUST00000039720.4
ENSMUST00000144175.2 |
Rragb
|
Ras-related GTP binding B |
| chr17_-_28441718 | 0.67 |
ENSMUST00000153744.1
|
Fkbp5
|
FK506 binding protein 5 |
| chr10_-_127341583 | 0.67 |
ENSMUST00000026474.3
|
Gli1
|
GLI-Kruppel family member GLI1 |
| chr17_-_10320229 | 0.66 |
ENSMUST00000053066.6
|
Qk
|
quaking |
| chr15_+_80133114 | 0.66 |
ENSMUST00000023050.7
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr3_+_103860265 | 0.66 |
ENSMUST00000029433.7
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr19_-_43752924 | 0.66 |
ENSMUST00000045562.5
|
Cox15
|
cytochrome c oxidase assembly protein 15 |
| chr2_+_28496891 | 0.66 |
ENSMUST00000163121.1
|
Gbgt1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 |
| chr2_+_152626932 | 0.65 |
ENSMUST00000000369.3
ENSMUST00000150913.1 |
Rem1
|
rad and gem related GTP binding protein 1 |
| chr5_-_74068361 | 0.62 |
ENSMUST00000119154.1
ENSMUST00000068058.7 |
Usp46
|
ubiquitin specific peptidase 46 |
| chr9_-_60687459 | 0.60 |
ENSMUST00000114032.1
ENSMUST00000166168.1 ENSMUST00000132366.1 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr18_+_65581704 | 0.59 |
ENSMUST00000182979.1
|
Zfp532
|
zinc finger protein 532 |
| chr16_+_17208135 | 0.59 |
ENSMUST00000169803.1
|
Rimbp3
|
RIMS binding protein 3 |
| chr5_-_140649018 | 0.58 |
ENSMUST00000042661.3
|
Ttyh3
|
tweety homolog 3 (Drosophila) |
| chr5_+_143651222 | 0.58 |
ENSMUST00000110727.1
|
Cyth3
|
cytohesin 3 |
| chr9_-_107635330 | 0.58 |
ENSMUST00000055704.6
|
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr3_-_19628669 | 0.58 |
ENSMUST00000119133.1
|
1700064H15Rik
|
RIKEN cDNA 1700064H15 gene |
| chr19_+_53140430 | 0.57 |
ENSMUST00000111741.2
|
Add3
|
adducin 3 (gamma) |
| chr13_-_112580662 | 0.57 |
ENSMUST00000051756.6
|
Il31ra
|
interleukin 31 receptor A |
| chr10_+_80930071 | 0.57 |
ENSMUST00000015456.8
|
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr16_+_23290464 | 0.57 |
ENSMUST00000115335.1
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1 |
| chrX_+_52791179 | 0.56 |
ENSMUST00000101588.1
|
Ccdc160
|
coiled-coil domain containing 160 |
| chr5_-_92278155 | 0.56 |
ENSMUST00000159345.1
ENSMUST00000113102.3 |
Naaa
|
N-acylethanolamine acid amidase |
| chr6_+_123229843 | 0.56 |
ENSMUST00000112554.2
ENSMUST00000024118.4 ENSMUST00000117130.1 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr11_+_3514861 | 0.54 |
ENSMUST00000094469.4
|
Selm
|
selenoprotein M |
| chr11_+_96464649 | 0.54 |
ENSMUST00000107663.3
|
Skap1
|
src family associated phosphoprotein 1 |
| chr2_+_24385313 | 0.54 |
ENSMUST00000056641.8
|
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr11_-_116412965 | 0.54 |
ENSMUST00000100202.3
ENSMUST00000106398.2 |
Rnf157
|
ring finger protein 157 |
| chr4_+_12906838 | 0.54 |
ENSMUST00000143186.1
ENSMUST00000183345.1 |
Triqk
|
triple QxxK/R motif containing |
| chr1_-_152625212 | 0.53 |
ENSMUST00000027760.7
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr6_+_86404257 | 0.53 |
ENSMUST00000095752.2
ENSMUST00000130967.1 |
Tia1
|
cytotoxic granule-associated RNA binding protein 1 |
| chr16_-_63864114 | 0.52 |
ENSMUST00000064405.6
|
Epha3
|
Eph receptor A3 |
| chr7_+_88430257 | 0.50 |
ENSMUST00000107256.2
|
Rab38
|
RAB38, member of RAS oncogene family |
| chr10_+_70204675 | 0.50 |
ENSMUST00000020090.1
|
2310015B20Rik
|
RIKEN cDNA 2310015B20 gene |
| chr8_+_21055047 | 0.50 |
ENSMUST00000098899.3
|
Defa23
|
defensin, alpha, 23 |
| chr4_+_155790439 | 0.50 |
ENSMUST00000165000.1
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr6_+_122921805 | 0.50 |
ENSMUST00000060484.8
|
Clec4a1
|
C-type lectin domain family 4, member a1 |
| chr7_-_133123770 | 0.49 |
ENSMUST00000164896.1
ENSMUST00000171968.1 |
Ctbp2
|
C-terminal binding protein 2 |
| chr7_-_46795661 | 0.49 |
ENSMUST00000123725.1
|
Hps5
|
Hermansky-Pudlak syndrome 5 homolog (human) |
| chr1_-_155232710 | 0.49 |
ENSMUST00000035914.3
|
BC034090
|
cDNA sequence BC034090 |
| chr11_-_75439551 | 0.48 |
ENSMUST00000128330.1
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr17_+_25016068 | 0.48 |
ENSMUST00000137386.1
|
Ift140
|
intraflagellar transport 140 |
| chr14_+_32833955 | 0.48 |
ENSMUST00000104926.2
|
Fam170b
|
family with sequence similarity 170, member B |
| chr5_+_117120120 | 0.48 |
ENSMUST00000111978.1
|
Taok3
|
TAO kinase 3 |
| chr7_-_44986313 | 0.48 |
ENSMUST00000045325.6
ENSMUST00000085387.4 ENSMUST00000107840.1 ENSMUST00000107843.3 ENSMUST00000107842.3 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr4_+_48585193 | 0.47 |
ENSMUST00000107703.1
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr19_-_53371766 | 0.47 |
ENSMUST00000086887.1
|
Gm10197
|
predicted gene 10197 |
| chr17_+_87635974 | 0.47 |
ENSMUST00000053577.8
|
Epcam
|
epithelial cell adhesion molecule |
| chr2_+_125247190 | 0.46 |
ENSMUST00000082122.7
|
Dut
|
deoxyuridine triphosphatase |
| chr11_+_60140066 | 0.46 |
ENSMUST00000171108.1
ENSMUST00000090806.4 |
Rai1
|
retinoic acid induced 1 |
| chr6_+_86404219 | 0.46 |
ENSMUST00000095754.3
ENSMUST00000095753.2 |
Tia1
|
cytotoxic granule-associated RNA binding protein 1 |
| chr19_-_6128208 | 0.46 |
ENSMUST00000025702.7
|
Snx15
|
sorting nexin 15 |
| chr6_+_86404336 | 0.45 |
ENSMUST00000113713.2
ENSMUST00000113708.1 |
Tia1
|
cytotoxic granule-associated RNA binding protein 1 |
| chr12_+_85599047 | 0.45 |
ENSMUST00000177587.1
|
Jdp2
|
Jun dimerization protein 2 |
| chr19_-_5366285 | 0.44 |
ENSMUST00000170010.1
|
Banf1
|
barrier to autointegration factor 1 |
| chr4_+_84884418 | 0.43 |
ENSMUST00000169371.2
|
Cntln
|
centlein, centrosomal protein |
| chr7_-_140082246 | 0.43 |
ENSMUST00000166758.2
|
Caly
|
calcyon neuron-specific vesicular protein |
| chr9_+_45370185 | 0.42 |
ENSMUST00000085939.6
|
Fxyd6
|
FXYD domain-containing ion transport regulator 6 |
| chr14_+_56402656 | 0.42 |
ENSMUST00000095793.1
|
Rnf17
|
ring finger protein 17 |
| chr7_-_4532419 | 0.41 |
ENSMUST00000094897.4
|
Dnaaf3
|
dynein, axonemal assembly factor 3 |
| chr7_-_119479249 | 0.41 |
ENSMUST00000033263.4
|
Umod
|
uromodulin |
| chr19_+_29951808 | 0.41 |
ENSMUST00000136850.1
|
Il33
|
interleukin 33 |
| chr10_-_87008015 | 0.41 |
ENSMUST00000035288.8
|
Stab2
|
stabilin 2 |
| chrX_-_102189371 | 0.41 |
ENSMUST00000033683.7
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr10_+_80855275 | 0.40 |
ENSMUST00000035597.8
|
Sppl2b
|
signal peptide peptidase like 2B |
| chr1_-_87101590 | 0.40 |
ENSMUST00000113270.2
|
Alpi
|
alkaline phosphatase, intestinal |
| chr1_+_178187417 | 0.40 |
ENSMUST00000161075.1
ENSMUST00000027783.7 |
Desi2
|
desumoylating isopeptidase 2 |
| chr12_+_79130777 | 0.40 |
ENSMUST00000021550.6
|
Arg2
|
arginase type II |
| chr19_-_6128144 | 0.39 |
ENSMUST00000154601.1
ENSMUST00000138931.1 |
Snx15
|
sorting nexin 15 |
| chr19_+_5024006 | 0.38 |
ENSMUST00000025826.5
|
Slc29a2
|
solute carrier family 29 (nucleoside transporters), member 2 |
| chr12_-_80260356 | 0.36 |
ENSMUST00000021554.8
|
Actn1
|
actinin, alpha 1 |
| chr6_-_69400097 | 0.36 |
ENSMUST00000177795.1
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr4_-_108406676 | 0.36 |
ENSMUST00000184609.1
|
Gpx7
|
glutathione peroxidase 7 |
| chr11_-_8973266 | 0.35 |
ENSMUST00000154153.1
|
Pkd1l1
|
polycystic kidney disease 1 like 1 |
| chr13_+_12395362 | 0.35 |
ENSMUST00000059270.8
|
Heatr1
|
HEAT repeat containing 1 |
| chr12_+_81631369 | 0.35 |
ENSMUST00000036116.5
|
Ttc9
|
tetratricopeptide repeat domain 9 |
| chr4_+_11558914 | 0.34 |
ENSMUST00000178703.1
ENSMUST00000095145.5 ENSMUST00000108306.2 ENSMUST00000070755.6 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr18_+_69925466 | 0.34 |
ENSMUST00000043929.4
|
Ccdc68
|
coiled-coil domain containing 68 |
| chrX_+_6873484 | 0.34 |
ENSMUST00000145302.1
|
Dgkk
|
diacylglycerol kinase kappa |
| chr6_-_34977999 | 0.33 |
ENSMUST00000044387.7
|
2010107G12Rik
|
RIKEN cDNA 2010107G12 gene |
| chr3_+_138352378 | 0.33 |
ENSMUST00000090166.4
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr15_+_82252397 | 0.33 |
ENSMUST00000136948.1
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr6_-_66559708 | 0.33 |
ENSMUST00000079584.1
|
Vmn1r32
|
vomeronasal 1 receptor 32 |
| chr5_-_149053038 | 0.33 |
ENSMUST00000085546.6
|
Hmgb1
|
high mobility group box 1 |
| chr17_-_35164891 | 0.32 |
ENSMUST00000025253.5
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr11_+_115475645 | 0.32 |
ENSMUST00000035240.6
|
Armc7
|
armadillo repeat containing 7 |
| chr11_-_40733373 | 0.31 |
ENSMUST00000020579.8
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr1_-_126738167 | 0.31 |
ENSMUST00000160693.1
|
Nckap5
|
NCK-associated protein 5 |
| chr18_-_60848911 | 0.31 |
ENSMUST00000177172.1
ENSMUST00000175934.1 ENSMUST00000176630.1 |
Tcof1
|
Treacher Collins Franceschetti syndrome 1, homolog |
| chr1_+_85575676 | 0.31 |
ENSMUST00000178024.1
|
G530012D18Rik
|
RIKEN cDNA G530012D1 gene |
| chr9_+_44334685 | 0.30 |
ENSMUST00000052686.2
|
H2afx
|
H2A histone family, member X |
| chr2_+_30077684 | 0.30 |
ENSMUST00000125346.1
|
Pkn3
|
protein kinase N3 |
| chr2_+_144033059 | 0.30 |
ENSMUST00000037722.2
ENSMUST00000110032.1 |
Banf2
|
barrier to autointegration factor 2 |
| chr19_+_43752996 | 0.30 |
ENSMUST00000026199.7
ENSMUST00000112047.3 ENSMUST00000153295.1 |
Cutc
|
cutC copper transporter homolog (E.coli) |
| chr5_-_92310003 | 0.30 |
ENSMUST00000031364.1
|
Sdad1
|
SDA1 domain containing 1 |
| chr5_+_48242549 | 0.30 |
ENSMUST00000172493.1
|
Slit2
|
slit homolog 2 (Drosophila) |
| chr14_-_105177280 | 0.29 |
ENSMUST00000100327.3
ENSMUST00000022715.7 |
Rbm26
|
RNA binding motif protein 26 |
| chr11_+_101246960 | 0.29 |
ENSMUST00000107282.3
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr12_-_111980751 | 0.28 |
ENSMUST00000170525.1
|
BC048943
|
cDNA sequence BC048943 |
| chr1_+_74153981 | 0.28 |
ENSMUST00000027372.7
ENSMUST00000106899.2 |
Cxcr2
|
chemokine (C-X-C motif) receptor 2 |
| chr11_-_116694802 | 0.28 |
ENSMUST00000079545.5
|
St6galnac2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
| chr18_+_42511496 | 0.28 |
ENSMUST00000025375.7
|
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr5_-_137613759 | 0.28 |
ENSMUST00000155251.1
ENSMUST00000124693.1 |
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr11_-_69837781 | 0.28 |
ENSMUST00000108634.2
|
Nlgn2
|
neuroligin 2 |
| chr11_+_70505244 | 0.28 |
ENSMUST00000019063.2
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr3_+_87796938 | 0.28 |
ENSMUST00000029711.2
ENSMUST00000107582.2 |
Insrr
|
insulin receptor-related receptor |
| chr4_-_141053660 | 0.28 |
ENSMUST00000040222.7
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr2_+_91922178 | 0.27 |
ENSMUST00000170432.1
|
Chrm4
|
cholinergic receptor, muscarinic 4 |
| chr17_+_29032664 | 0.27 |
ENSMUST00000130216.1
|
Srsf3
|
serine/arginine-rich splicing factor 3 |
| chr11_-_86257553 | 0.27 |
ENSMUST00000132024.1
ENSMUST00000139285.1 |
Ints2
|
integrator complex subunit 2 |
| chrX_-_139714481 | 0.27 |
ENSMUST00000183728.1
|
Gm15013
|
predicted gene 15013 |
| chr5_+_12383156 | 0.27 |
ENSMUST00000030868.6
|
Sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr2_+_181680284 | 0.26 |
ENSMUST00000103042.3
|
Tcea2
|
transcription elongation factor A (SII), 2 |
| chr4_+_84884276 | 0.26 |
ENSMUST00000047023.6
|
Cntln
|
centlein, centrosomal protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Crx_Gsc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0009753 | sesquiterpenoid metabolic process(GO:0006714) response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.7 | 0.7 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.6 | 4.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 2.6 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.4 | 1.2 | GO:0070315 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.4 | 2.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.4 | 1.9 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) protein secretion by platelet(GO:0070560) |
| 0.4 | 3.5 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.3 | 3.3 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.3 | 5.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.3 | 3.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.3 | 2.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 1.9 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 3.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.0 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.2 | 1.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.2 | 0.7 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.2 | 1.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.2 | 0.5 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.2 | 0.5 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.2 | 2.9 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.2 | 2.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 0.9 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 3.4 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.2 | 0.7 | GO:0060032 | notochord regression(GO:0060032) |
| 0.2 | 0.7 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.0 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.2 | 0.5 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 0.1 | 0.9 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 1.5 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 0.3 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.1 | 0.8 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.8 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.5 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 0.5 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.7 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.4 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.1 | 0.5 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.3 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.1 | 0.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.4 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.1 | 0.7 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.4 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.1 | 0.7 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.1 | 0.3 | GO:0050929 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 | 0.5 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 1.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 1.0 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 0.2 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.1 | 0.3 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.4 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 1.7 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.1 | 0.7 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.2 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.1 | 0.3 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 0.6 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.3 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.6 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.2 | GO:0072554 | Notch signaling pathway involved in heart induction(GO:0003137) regulation of Notch signaling pathway involved in heart induction(GO:0035480) positive regulation of Notch signaling pathway involved in heart induction(GO:0035481) arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.2 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.7 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.2 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 0.5 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 1.0 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.1 | 0.3 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.9 | GO:0009437 | carnitine metabolic process(GO:0009437) positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 1.0 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.1 | 0.2 | GO:0090273 | somatostatin secretion(GO:0070253) regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 0.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.0 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.6 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.8 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 1.2 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 0.7 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.4 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 1.1 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0032240 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.5 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.5 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 1.5 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.8 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.2 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.4 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.1 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.5 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.4 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.7 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.4 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.3 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) B cell homeostatic proliferation(GO:0002358) |
| 0.0 | 1.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.1 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.0 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.0 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.4 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.3 | 0.9 | GO:0000799 | nuclear condensin complex(GO:0000799) germinal vesicle(GO:0042585) |
| 0.2 | 3.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 0.8 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 1.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 2.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.7 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.4 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.1 | 0.5 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 6.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 4.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.6 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.4 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.4 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 1.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.7 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 0.6 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.6 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.7 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 4.5 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 2.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.0 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 7.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.7 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 9.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.1 | 4.5 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 1.0 | 3.1 | GO:0047787 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.9 | 2.6 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.6 | 2.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 6.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 3.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 1.1 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.3 | 3.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 3.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 1.9 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 3.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 0.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 0.5 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.2 | 0.5 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 2.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 3.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.3 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.1 | 0.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.6 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.7 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 2.0 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 1.0 | GO:0070679 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.5 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 2.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.5 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 1.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.2 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 1.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 2.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 5.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.0 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.7 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 6.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 3.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 3.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 3.8 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 3.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 5.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 11.7 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 0.6 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.2 | 3.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 0.7 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.1 | 0.7 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 4.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.8 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.1 | 0.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 2.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.8 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 2.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |