Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
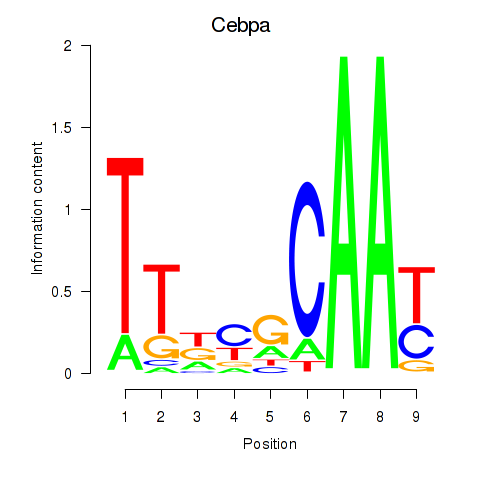
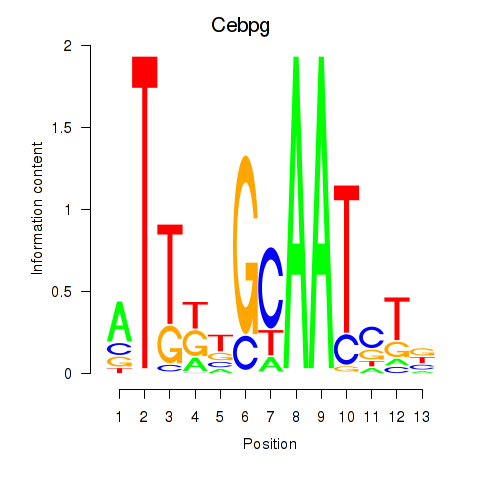
Results for Cebpa_Cebpg
Z-value: 1.08
Transcription factors associated with Cebpa_Cebpg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cebpa
|
ENSMUSG00000034957.9 | CCAAT/enhancer binding protein (C/EBP), alpha |
|
Cebpg
|
ENSMUSG00000056216.8 | CCAAT/enhancer binding protein (C/EBP), gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cebpa | mm10_v2_chr7_+_35119285_35119301 | 0.58 | 2.1e-04 | Click! |
| Cebpg | mm10_v2_chr7_-_35056467_35056556 | 0.06 | 7.2e-01 | Click! |
Activity profile of Cebpa_Cebpg motif
Sorted Z-values of Cebpa_Cebpg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_13623967 | 16.89 |
ENSMUST00000108525.2
|
Sult2a5
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 5 |
| chr7_+_13733502 | 13.71 |
ENSMUST00000086148.6
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr7_-_14123042 | 10.67 |
ENSMUST00000098809.2
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr7_-_46742979 | 6.69 |
ENSMUST00000128088.1
|
Saa1
|
serum amyloid A 1 |
| chr7_-_13989588 | 6.60 |
ENSMUST00000165167.1
ENSMUST00000108520.2 |
Sult2a4
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 4 |
| chr2_+_173153048 | 6.36 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr7_+_46751832 | 5.67 |
ENSMUST00000075982.2
|
Saa2
|
serum amyloid A 2 |
| chr3_+_19957088 | 5.55 |
ENSMUST00000108328.1
|
Cp
|
ceruloplasmin |
| chr3_+_19957037 | 5.37 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr7_-_14254870 | 5.35 |
ENSMUST00000184731.1
ENSMUST00000076576.6 |
Sult2a6
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 6 |
| chr19_-_7802578 | 5.08 |
ENSMUST00000120522.1
ENSMUST00000065634.7 |
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr3_+_19957240 | 4.93 |
ENSMUST00000108325.2
|
Cp
|
ceruloplasmin |
| chr7_+_26061495 | 4.50 |
ENSMUST00000005669.7
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr16_+_22892035 | 3.75 |
ENSMUST00000023583.5
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr5_+_90561102 | 3.70 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr10_+_23851454 | 3.51 |
ENSMUST00000020190.7
|
Vnn3
|
vanin 3 |
| chr14_+_51091077 | 3.18 |
ENSMUST00000022428.5
ENSMUST00000171688.1 |
Rnase4
Ang
|
ribonuclease, RNase A family 4 angiogenin, ribonuclease, RNase A family, 5 |
| chr14_+_65970610 | 3.11 |
ENSMUST00000127387.1
|
Clu
|
clusterin |
| chr8_-_93131271 | 2.93 |
ENSMUST00000034189.8
|
Ces1c
|
carboxylesterase 1C |
| chr14_+_65971049 | 2.88 |
ENSMUST00000128539.1
|
Clu
|
clusterin |
| chr12_-_80968075 | 2.85 |
ENSMUST00000095572.4
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr14_+_65971164 | 2.59 |
ENSMUST00000144619.1
|
Clu
|
clusterin |
| chr16_+_23058250 | 2.41 |
ENSMUST00000039492.6
ENSMUST00000023589.8 ENSMUST00000089902.6 |
Kng1
|
kininogen 1 |
| chr1_+_130865669 | 2.40 |
ENSMUST00000038829.5
|
Faim3
|
Fas apoptotic inhibitory molecule 3 |
| chr17_-_56121946 | 2.30 |
ENSMUST00000041357.7
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr6_-_124542281 | 2.28 |
ENSMUST00000159463.1
ENSMUST00000162844.1 ENSMUST00000160505.1 ENSMUST00000162443.1 |
C1s
|
complement component 1, s subcomponent |
| chr10_-_102490418 | 2.25 |
ENSMUST00000020040.3
|
Nts
|
neurotensin |
| chr16_-_23029062 | 2.14 |
ENSMUST00000115349.2
|
Kng2
|
kininogen 2 |
| chr16_-_23029012 | 2.13 |
ENSMUST00000039338.6
|
Kng2
|
kininogen 2 |
| chr16_-_23029080 | 2.13 |
ENSMUST00000100046.2
|
Kng2
|
kininogen 2 |
| chr7_-_45615484 | 2.12 |
ENSMUST00000033099.4
|
Fgf21
|
fibroblast growth factor 21 |
| chr14_+_65970804 | 2.07 |
ENSMUST00000138191.1
|
Clu
|
clusterin |
| chr2_-_32424005 | 2.01 |
ENSMUST00000113307.2
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr1_+_169655493 | 1.98 |
ENSMUST00000027997.3
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr3_-_89913144 | 1.97 |
ENSMUST00000029559.6
|
Il6ra
|
interleukin 6 receptor, alpha |
| chr6_+_141629499 | 1.89 |
ENSMUST00000042812.6
|
Slco1b2
|
solute carrier organic anion transporter family, member 1b2 |
| chr11_-_99493112 | 1.83 |
ENSMUST00000006969.7
|
Krt23
|
keratin 23 |
| chr2_-_129371131 | 1.82 |
ENSMUST00000028881.7
|
Il1b
|
interleukin 1 beta |
| chr17_-_57228003 | 1.78 |
ENSMUST00000177046.1
ENSMUST00000024988.8 |
C3
|
complement component 3 |
| chr19_+_18749983 | 1.71 |
ENSMUST00000040489.7
|
Trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr16_-_93929512 | 1.64 |
ENSMUST00000177648.1
|
Cldn14
|
claudin 14 |
| chr2_+_102706356 | 1.50 |
ENSMUST00000123759.1
ENSMUST00000111212.1 ENSMUST00000005220.4 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chrX_-_147429189 | 1.44 |
ENSMUST00000033646.2
|
Il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr14_-_45477856 | 1.41 |
ENSMUST00000141424.1
|
Fermt2
|
fermitin family homolog 2 (Drosophila) |
| chr11_+_4031770 | 1.37 |
ENSMUST00000019512.7
|
Sec14l4
|
SEC14-like 4 (S. cerevisiae) |
| chr11_+_67200052 | 1.37 |
ENSMUST00000124516.1
ENSMUST00000018637.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr7_+_127800604 | 1.36 |
ENSMUST00000046863.5
ENSMUST00000106272.1 ENSMUST00000139068.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr1_+_164796723 | 1.33 |
ENSMUST00000027861.4
|
Dpt
|
dermatopontin |
| chr1_-_71653162 | 1.29 |
ENSMUST00000055226.6
|
Fn1
|
fibronectin 1 |
| chrX_-_162964557 | 1.27 |
ENSMUST00000038769.2
|
S100g
|
S100 calcium binding protein G |
| chr6_+_17463749 | 1.21 |
ENSMUST00000115443.1
|
Met
|
met proto-oncogene |
| chr4_+_63362443 | 1.19 |
ENSMUST00000075341.3
|
Orm2
|
orosomucoid 2 |
| chr15_-_3582596 | 1.10 |
ENSMUST00000161770.1
|
Ghr
|
growth hormone receptor |
| chr9_+_108080436 | 1.10 |
ENSMUST00000035211.7
ENSMUST00000162886.1 |
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr7_+_26307190 | 1.08 |
ENSMUST00000098657.3
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr15_+_87625214 | 1.08 |
ENSMUST00000068088.6
|
Fam19a5
|
family with sequence similarity 19, member A5 |
| chr2_-_160872985 | 1.05 |
ENSMUST00000109460.1
ENSMUST00000127201.1 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr12_+_100779055 | 1.05 |
ENSMUST00000069782.4
|
9030617O03Rik
|
RIKEN cDNA 9030617O03 gene |
| chr17_-_13404191 | 1.04 |
ENSMUST00000115650.1
|
Gm8597
|
predicted gene 8597 |
| chr12_+_100779074 | 1.00 |
ENSMUST00000110073.1
ENSMUST00000110070.1 |
9030617O03Rik
|
RIKEN cDNA 9030617O03 gene |
| chr14_-_18893376 | 1.00 |
ENSMUST00000151926.1
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr12_+_100779088 | 0.97 |
ENSMUST00000110069.1
|
9030617O03Rik
|
RIKEN cDNA 9030617O03 gene |
| chr18_+_20944607 | 0.94 |
ENSMUST00000050004.1
|
Rnf125
|
ring finger protein 125 |
| chr19_+_34290653 | 0.94 |
ENSMUST00000025691.5
ENSMUST00000112472.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr3_-_107986360 | 0.94 |
ENSMUST00000066530.6
|
Gstm2
|
glutathione S-transferase, mu 2 |
| chr3_-_107986408 | 0.93 |
ENSMUST00000012348.2
|
Gstm2
|
glutathione S-transferase, mu 2 |
| chrX_+_59999436 | 0.92 |
ENSMUST00000033477.4
|
F9
|
coagulation factor IX |
| chr10_+_93488766 | 0.89 |
ENSMUST00000129421.1
|
Hal
|
histidine ammonia lyase |
| chr7_-_127273919 | 0.88 |
ENSMUST00000082428.3
|
Sephs2
|
selenophosphate synthetase 2 |
| chr8_-_86580664 | 0.87 |
ENSMUST00000131423.1
ENSMUST00000152438.1 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr2_-_160872829 | 0.84 |
ENSMUST00000176141.1
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr11_-_110095937 | 0.82 |
ENSMUST00000106664.3
ENSMUST00000046223.7 ENSMUST00000106662.1 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr16_-_38433145 | 0.82 |
ENSMUST00000002926.6
|
Pla1a
|
phospholipase A1 member A |
| chr11_+_108395288 | 0.79 |
ENSMUST00000000049.5
|
Apoh
|
apolipoprotein H |
| chr10_+_21377290 | 0.79 |
ENSMUST00000042699.7
ENSMUST00000159163.1 |
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr2_-_52676571 | 0.77 |
ENSMUST00000178799.1
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr2_-_164743182 | 0.77 |
ENSMUST00000103096.3
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr14_+_75242287 | 0.76 |
ENSMUST00000022576.8
|
Cpb2
|
carboxypeptidase B2 (plasma) |
| chr9_+_7571396 | 0.71 |
ENSMUST00000120900.1
ENSMUST00000093896.3 ENSMUST00000151853.1 ENSMUST00000152878.1 |
Mmp27
|
matrix metallopeptidase 27 |
| chr9_+_94669876 | 0.70 |
ENSMUST00000033463.9
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr8_-_46211284 | 0.70 |
ENSMUST00000034049.4
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chr5_-_103977326 | 0.69 |
ENSMUST00000120320.1
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr17_+_28858411 | 0.69 |
ENSMUST00000114737.1
ENSMUST00000056866.5 |
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr13_+_24943144 | 0.68 |
ENSMUST00000021773.5
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr4_-_155361322 | 0.68 |
ENSMUST00000105624.1
|
Prkcz
|
protein kinase C, zeta |
| chr5_-_103977360 | 0.66 |
ENSMUST00000048118.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr16_-_30267524 | 0.65 |
ENSMUST00000064856.7
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr5_-_103977404 | 0.65 |
ENSMUST00000112803.2
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_-_51567717 | 0.64 |
ENSMUST00000127135.1
ENSMUST00000151104.1 |
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr2_-_65567505 | 0.62 |
ENSMUST00000100069.2
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr15_+_100304782 | 0.61 |
ENSMUST00000067752.3
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr3_-_151835967 | 0.61 |
ENSMUST00000106126.1
|
Ptgfr
|
prostaglandin F receptor |
| chr19_-_36119833 | 0.56 |
ENSMUST00000025718.8
|
Ankrd1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr18_-_35498856 | 0.55 |
ENSMUST00000025215.8
|
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr13_-_78199757 | 0.55 |
ENSMUST00000091458.6
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chrX_+_8874253 | 0.54 |
ENSMUST00000115561.1
|
Gm5751
|
predicted gene 5751 |
| chr14_-_43923559 | 0.54 |
ENSMUST00000159175.1
|
Ear10
|
eosinophil-associated, ribonuclease A family, member 10 |
| chr3_-_75270073 | 0.54 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr15_+_31224371 | 0.54 |
ENSMUST00000044524.9
|
Dap
|
death-associated protein |
| chr14_-_43923368 | 0.53 |
ENSMUST00000163652.1
|
Ear10
|
eosinophil-associated, ribonuclease A family, member 10 |
| chr8_-_41054771 | 0.51 |
ENSMUST00000093534.4
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr4_+_155562348 | 0.51 |
ENSMUST00000030939.7
|
Nadk
|
NAD kinase |
| chr5_-_99252839 | 0.50 |
ENSMUST00000168092.1
ENSMUST00000031276.8 |
Rasgef1b
|
RasGEF domain family, member 1B |
| chr2_-_65567465 | 0.48 |
ENSMUST00000066432.5
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr5_+_102724971 | 0.48 |
ENSMUST00000112853.1
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr12_-_111377705 | 0.47 |
ENSMUST00000041965.3
|
Cdc42bpb
|
CDC42 binding protein kinase beta |
| chr16_-_34095983 | 0.47 |
ENSMUST00000114973.1
ENSMUST00000114964.1 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr2_-_104849465 | 0.45 |
ENSMUST00000126824.1
|
Prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr5_+_63812447 | 0.44 |
ENSMUST00000081747.3
|
0610040J01Rik
|
RIKEN cDNA 0610040J01 gene |
| chrX_+_94636066 | 0.44 |
ENSMUST00000096368.3
|
Gspt2
|
G1 to S phase transition 2 |
| chr11_-_106920359 | 0.40 |
ENSMUST00000167787.1
ENSMUST00000092517.2 |
Smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr15_+_54952939 | 0.40 |
ENSMUST00000181704.1
|
Gm26684
|
predicted gene, 26684 |
| chr19_-_34877880 | 0.39 |
ENSMUST00000112460.1
|
Pank1
|
pantothenate kinase 1 |
| chr3_-_104366613 | 0.37 |
ENSMUST00000056145.2
|
Gm5546
|
predicted gene 5546 |
| chr16_-_13903051 | 0.37 |
ENSMUST00000115803.1
|
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr11_+_75732869 | 0.36 |
ENSMUST00000067664.3
|
Ywhae
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
| chr14_+_22019833 | 0.35 |
ENSMUST00000159777.1
ENSMUST00000162540.1 |
1700112E06Rik
|
RIKEN cDNA 1700112E06 gene |
| chr8_+_22227322 | 0.35 |
ENSMUST00000160585.1
|
Thsd1
|
thrombospondin, type I, domain 1 |
| chr15_-_65014904 | 0.34 |
ENSMUST00000110100.2
|
Gm21961
|
predicted gene, 21961 |
| chr5_+_30013141 | 0.34 |
ENSMUST00000026845.7
|
Il6
|
interleukin 6 |
| chr7_-_100371889 | 0.33 |
ENSMUST00000032963.8
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr13_-_21292679 | 0.33 |
ENSMUST00000004456.4
ENSMUST00000110491.1 |
Gpx5
|
glutathione peroxidase 5 |
| chr11_-_43901187 | 0.31 |
ENSMUST00000067258.2
ENSMUST00000139906.1 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr4_-_63861326 | 0.31 |
ENSMUST00000030047.2
|
Tnfsf8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr10_+_80930071 | 0.31 |
ENSMUST00000015456.8
|
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr4_+_136143497 | 0.31 |
ENSMUST00000008016.2
|
Id3
|
inhibitor of DNA binding 3 |
| chr7_+_51880312 | 0.29 |
ENSMUST00000145049.1
|
Gas2
|
growth arrest specific 2 |
| chr14_+_44102654 | 0.28 |
ENSMUST00000074839.6
|
Ear2
|
eosinophil-associated, ribonuclease A family, member 2 |
| chr16_-_13903085 | 0.28 |
ENSMUST00000023361.5
ENSMUST00000115802.1 |
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr15_-_55906722 | 0.27 |
ENSMUST00000110200.2
|
Sntb1
|
syntrophin, basic 1 |
| chr18_-_52529847 | 0.27 |
ENSMUST00000171470.1
|
Lox
|
lysyl oxidase |
| chr17_+_36942910 | 0.26 |
ENSMUST00000040498.5
|
Rnf39
|
ring finger protein 39 |
| chr3_-_83049797 | 0.26 |
ENSMUST00000048246.3
|
Fgb
|
fibrinogen beta chain |
| chr16_-_13903015 | 0.25 |
ENSMUST00000115804.2
|
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr18_-_52529692 | 0.25 |
ENSMUST00000025409.7
|
Lox
|
lysyl oxidase |
| chr14_+_51203689 | 0.25 |
ENSMUST00000049559.1
|
Ear14
|
eosinophil-associated, ribonuclease A family, member 14 |
| chr10_+_5593718 | 0.24 |
ENSMUST00000051809.8
|
Myct1
|
myc target 1 |
| chr3_-_63964659 | 0.24 |
ENSMUST00000161659.1
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr1_-_39577340 | 0.23 |
ENSMUST00000062525.5
|
Rnf149
|
ring finger protein 149 |
| chr17_+_36943025 | 0.23 |
ENSMUST00000173072.1
|
Rnf39
|
ring finger protein 39 |
| chr9_-_40006947 | 0.23 |
ENSMUST00000073932.3
|
Olfr980
|
olfactory receptor 980 |
| chr15_+_44619551 | 0.23 |
ENSMUST00000022964.7
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr13_+_31806627 | 0.22 |
ENSMUST00000062292.2
|
Foxc1
|
forkhead box C1 |
| chr4_-_132732514 | 0.20 |
ENSMUST00000045550.4
|
Xkr8
|
X Kell blood group precursor related family member 8 homolog |
| chr14_-_43875517 | 0.20 |
ENSMUST00000179200.1
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr11_+_98358368 | 0.20 |
ENSMUST00000018311.4
|
Stard3
|
START domain containing 3 |
| chr4_-_15149051 | 0.20 |
ENSMUST00000041606.7
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr11_-_84069179 | 0.19 |
ENSMUST00000138208.1
|
Dusp14
|
dual specificity phosphatase 14 |
| chr7_-_79149042 | 0.19 |
ENSMUST00000032825.8
ENSMUST00000107409.3 |
Mfge8
|
milk fat globule-EGF factor 8 protein |
| chr11_+_87853207 | 0.18 |
ENSMUST00000038196.6
|
Mks1
|
Meckel syndrome, type 1 |
| chr14_+_22019712 | 0.18 |
ENSMUST00000075639.4
ENSMUST00000161249.1 |
1700112E06Rik
|
RIKEN cDNA 1700112E06 gene |
| chr3_-_82876483 | 0.18 |
ENSMUST00000048647.7
|
Rbm46
|
RNA binding motif protein 46 |
| chr17_+_28691342 | 0.17 |
ENSMUST00000114758.1
ENSMUST00000004990.6 ENSMUST00000062694.8 ENSMUST00000114754.1 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr16_+_3847206 | 0.16 |
ENSMUST00000041778.7
|
Zfp174
|
zinc finger protein 174 |
| chr10_-_62651194 | 0.16 |
ENSMUST00000020270.4
|
Ddx50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr1_-_52490736 | 0.15 |
ENSMUST00000170269.1
|
Nab1
|
Ngfi-A binding protein 1 |
| chr6_-_67491849 | 0.15 |
ENSMUST00000118364.1
|
Il23r
|
interleukin 23 receptor |
| chr3_-_63964768 | 0.15 |
ENSMUST00000029402.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr18_+_37489465 | 0.15 |
ENSMUST00000055949.2
|
Pcdhb18
|
protocadherin beta 18 |
| chr14_-_43819639 | 0.15 |
ENSMUST00000100691.3
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr5_-_92083667 | 0.15 |
ENSMUST00000113127.3
|
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chrM_+_8600 | 0.14 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr11_+_59306920 | 0.13 |
ENSMUST00000000128.3
ENSMUST00000108783.3 |
Wnt9a
|
wingless-type MMTV integration site 9A |
| chr12_+_98268626 | 0.13 |
ENSMUST00000075072.4
|
Gpr65
|
G-protein coupled receptor 65 |
| chr2_-_144011202 | 0.13 |
ENSMUST00000016072.5
ENSMUST00000037875.5 |
Rrbp1
|
ribosome binding protein 1 |
| chr11_+_70525361 | 0.12 |
ENSMUST00000018430.6
|
Psmb6
|
proteasome (prosome, macropain) subunit, beta type 6 |
| chr11_+_78037959 | 0.11 |
ENSMUST00000073660.6
|
Flot2
|
flotillin 2 |
| chr6_-_40436104 | 0.11 |
ENSMUST00000039008.6
ENSMUST00000101492.3 |
E330009J07Rik
|
RIKEN cDNA E330009J07 gene |
| chr5_-_91402905 | 0.11 |
ENSMUST00000121044.2
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr18_-_42899294 | 0.11 |
ENSMUST00000117687.1
|
Ppp2r2b
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform |
| chr7_+_3645267 | 0.10 |
ENSMUST00000038913.9
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr14_-_51071442 | 0.10 |
ENSMUST00000048478.5
|
Olfr750
|
olfactory receptor 750 |
| chr1_-_153851189 | 0.09 |
ENSMUST00000059607.6
|
5830403L16Rik
|
RIKEN cDNA 5830403L16 gene |
| chr1_-_132707304 | 0.09 |
ENSMUST00000043189.7
|
Nfasc
|
neurofascin |
| chr17_+_86963077 | 0.09 |
ENSMUST00000024956.8
|
Rhoq
|
ras homolog gene family, member Q |
| chr17_+_32284772 | 0.08 |
ENSMUST00000181112.1
|
Gm26549
|
predicted gene, 26549 |
| chr6_+_96115249 | 0.08 |
ENSMUST00000075080.5
|
Fam19a1
|
family with sequence similarity 19, member A1 |
| chr1_+_134494628 | 0.08 |
ENSMUST00000047978.7
|
Rabif
|
RAB interacting factor |
| chr13_+_51100810 | 0.07 |
ENSMUST00000095797.5
|
Spin1
|
spindlin 1 |
| chr18_-_34931993 | 0.07 |
ENSMUST00000025218.7
|
Etf1
|
eukaryotic translation termination factor 1 |
| chr15_-_58214882 | 0.06 |
ENSMUST00000022986.6
|
Fbxo32
|
F-box protein 32 |
| chr15_+_9140527 | 0.06 |
ENSMUST00000090380.4
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr9_+_5298517 | 0.06 |
ENSMUST00000027015.5
|
Casp1
|
caspase 1 |
| chr7_-_25754701 | 0.05 |
ENSMUST00000108401.1
ENSMUST00000043765.7 |
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr1_+_60343297 | 0.05 |
ENSMUST00000114202.1
ENSMUST00000060608.6 |
Cyp20a1
|
cytochrome P450, family 20, subfamily a, polypeptide 1 |
| chr5_-_137533170 | 0.05 |
ENSMUST00000168746.1
ENSMUST00000170293.1 |
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chrX_+_6047453 | 0.05 |
ENSMUST00000103007.3
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr16_+_42907563 | 0.05 |
ENSMUST00000151244.1
ENSMUST00000114694.2 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr7_+_103550368 | 0.05 |
ENSMUST00000106888.1
|
Olfr613
|
olfactory receptor 613 |
| chr2_+_104590453 | 0.04 |
ENSMUST00000028599.7
|
Cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr11_-_35798884 | 0.04 |
ENSMUST00000160726.2
|
Fbll1
|
fibrillarin-like 1 |
| chr12_+_86734381 | 0.04 |
ENSMUST00000095527.5
|
Gm6772
|
predicted gene 6772 |
| chr16_+_20097554 | 0.04 |
ENSMUST00000023509.3
|
Klhl24
|
kelch-like 24 |
| chr14_+_51853699 | 0.04 |
ENSMUST00000169070.1
ENSMUST00000074477.6 |
Ear6
|
eosinophil-associated, ribonuclease A family, member 6 |
| chrX_+_71972986 | 0.04 |
ENSMUST00000066116.1
|
Fate1
|
fetal and adult testis expressed 1 |
| chr17_+_28691419 | 0.03 |
ENSMUST00000124886.1
|
Mapk14
|
mitogen-activated protein kinase 14 |
| chr11_+_23020464 | 0.03 |
ENSMUST00000094363.3
ENSMUST00000151877.1 |
Fam161a
|
family with sequence similarity 161, member A |
| chr2_+_78051220 | 0.02 |
ENSMUST00000144728.1
|
4930440I19Rik
|
RIKEN cDNA 4930440I19 gene |
| chr12_-_105222734 | 0.02 |
ENSMUST00000041316.8
ENSMUST00000175652.1 |
Tcl1
|
T cell lymphoma breakpoint 1 |
| chr18_-_84681966 | 0.02 |
ENSMUST00000168419.1
|
Cndp2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cebpa_Cebpg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 1.8 | 10.6 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.3 | 1.3 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.8 | 3.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.7 | 0.7 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.6 | 1.8 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.5 | 2.0 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.5 | 15.8 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.4 | 0.8 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.4 | 5.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.4 | 1.8 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.3 | 15.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.3 | 1.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.3 | 1.9 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.3 | 2.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 1.2 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.2 | 0.7 | GO:0006507 | GPI anchor release(GO:0006507) response to iron(II) ion(GO:0010040) |
| 0.2 | 0.9 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.2 | 1.5 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 0.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 0.9 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.2 | 0.9 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.2 | 4.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 5.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 1.4 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.2 | 0.6 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.2 | 0.3 | GO:0045818 | negative regulation of glycogen catabolic process(GO:0045818) |
| 0.2 | 0.6 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.2 | 1.4 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 1.1 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 6.7 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.8 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 1.1 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 3.5 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 0.7 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.1 | 0.8 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.4 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 2.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.2 | GO:1990868 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.1 | 0.4 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 2.3 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.5 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.7 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 1.4 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 | 1.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 11.1 | GO:0008202 | steroid metabolic process(GO:0008202) |
| 0.0 | 2.2 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 1.7 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 2.0 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 | 0.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0030913 | protein localization to paranode region of axon(GO:0002175) paranodal junction assembly(GO:0030913) |
| 0.0 | 1.6 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.2 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.0 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.5 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.3 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.8 | 10.6 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.6 | 2.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 12.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 1.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.3 | 15.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 1.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.9 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 13.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 1.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 1.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 3.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 3.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 39.5 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 1.6 | 15.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.8 | 3.8 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.7 | 2.0 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.4 | 10.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 2.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 1.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.3 | 11.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 5.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 0.9 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 1.2 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.3 | 6.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 1.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 1.9 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.2 | 2.7 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 13.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.2 | 1.3 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.2 | 1.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 1.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 5.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.8 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.9 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 2.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.9 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 0.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 5.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 3.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.1 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.6 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 1.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 2.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 3.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 3.3 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 2.0 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.2 | GO:0005262 | calcium channel activity(GO:0005262) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 6.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 2.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 15.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 10.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 6.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 2.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.4 | 15.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.4 | 6.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 2.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 2.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 1.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 3.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 12.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |