Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
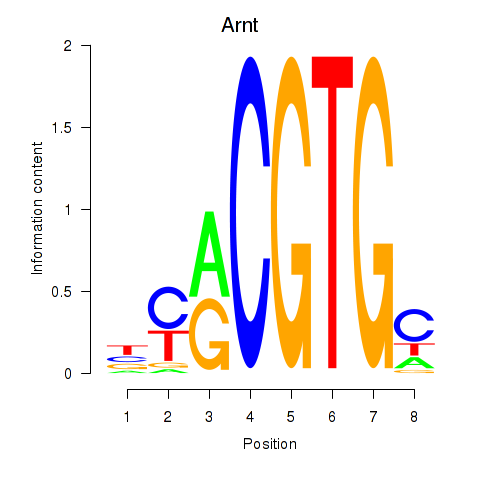
Results for Arnt
Z-value: 1.02
Transcription factors associated with Arnt
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arnt
|
ENSMUSG00000015522.12 | aryl hydrocarbon receptor nuclear translocator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arnt | mm10_v2_chr3_+_95434386_95434428 | -0.62 | 6.3e-05 | Click! |
Activity profile of Arnt motif
Sorted Z-values of Arnt motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_105269837 | 6.27 |
ENSMUST00000172525.1
ENSMUST00000174837.1 ENSMUST00000173859.1 |
Hsf4
|
heat shock transcription factor 4 |
| chr8_+_105269788 | 6.07 |
ENSMUST00000036127.2
ENSMUST00000163734.2 |
Hsf4
|
heat shock transcription factor 4 |
| chr17_+_36942910 | 5.55 |
ENSMUST00000040498.5
|
Rnf39
|
ring finger protein 39 |
| chr14_+_69171576 | 5.18 |
ENSMUST00000062437.8
|
Nkx2-6
|
NK2 homeobox 6 |
| chr1_-_121327672 | 4.64 |
ENSMUST00000159085.1
ENSMUST00000159125.1 ENSMUST00000161818.1 |
Insig2
|
insulin induced gene 2 |
| chr17_+_36943025 | 4.58 |
ENSMUST00000173072.1
|
Rnf39
|
ring finger protein 39 |
| chr1_-_121327734 | 4.38 |
ENSMUST00000160968.1
ENSMUST00000162582.1 |
Insig2
|
insulin induced gene 2 |
| chr1_-_121328024 | 4.33 |
ENSMUST00000003818.7
|
Insig2
|
insulin induced gene 2 |
| chr1_-_121327776 | 4.05 |
ENSMUST00000160688.1
|
Insig2
|
insulin induced gene 2 |
| chr19_+_7056731 | 3.35 |
ENSMUST00000040261.5
|
Macrod1
|
MACRO domain containing 1 |
| chr8_+_13159135 | 3.25 |
ENSMUST00000033824.6
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr6_+_64729118 | 3.19 |
ENSMUST00000101351.4
|
Atoh1
|
atonal homolog 1 (Drosophila) |
| chr3_-_95882031 | 3.09 |
ENSMUST00000161994.1
|
Gm129
|
predicted gene 129 |
| chr5_-_34187670 | 3.05 |
ENSMUST00000042701.6
ENSMUST00000119171.1 |
Mxd4
|
Max dimerization protein 4 |
| chr11_-_116199040 | 3.00 |
ENSMUST00000066587.5
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr3_-_95882193 | 2.95 |
ENSMUST00000159863.1
ENSMUST00000159739.1 ENSMUST00000036418.3 |
Gm129
|
predicted gene 129 |
| chr9_+_46012810 | 2.90 |
ENSMUST00000126865.1
|
Sik3
|
SIK family kinase 3 |
| chr11_-_98775333 | 2.86 |
ENSMUST00000064941.6
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr15_+_81811414 | 2.68 |
ENSMUST00000023024.7
|
Tef
|
thyrotroph embryonic factor |
| chr9_-_106685653 | 2.46 |
ENSMUST00000163441.1
|
Tex264
|
testis expressed gene 264 |
| chr5_-_65435717 | 2.45 |
ENSMUST00000117542.1
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr5_-_65435881 | 2.38 |
ENSMUST00000031103.7
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr3_-_95882232 | 2.38 |
ENSMUST00000161866.1
|
Gm129
|
predicted gene 129 |
| chr4_+_125490688 | 2.26 |
ENSMUST00000030676.7
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr4_+_97777780 | 2.23 |
ENSMUST00000107062.2
ENSMUST00000052018.5 ENSMUST00000107057.1 |
Nfia
|
nuclear factor I/A |
| chr9_+_77917364 | 2.23 |
ENSMUST00000034904.7
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr17_+_24752980 | 2.08 |
ENSMUST00000044922.6
|
Hs3st6
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
| chr14_-_18239053 | 2.01 |
ENSMUST00000090543.5
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr8_+_56294552 | 1.91 |
ENSMUST00000034026.8
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD) |
| chr19_-_42128982 | 1.90 |
ENSMUST00000161873.1
|
Avpi1
|
arginine vasopressin-induced 1 |
| chr1_-_179546261 | 1.86 |
ENSMUST00000027769.5
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr2_+_122234749 | 1.83 |
ENSMUST00000110551.3
|
Sord
|
sorbitol dehydrogenase |
| chr1_+_151344472 | 1.81 |
ENSMUST00000023918.6
ENSMUST00000097543.1 ENSMUST00000111887.3 |
Ivns1abp
|
influenza virus NS1A binding protein |
| chr11_+_120491840 | 1.80 |
ENSMUST00000026899.3
|
Slc25a10
|
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 |
| chr19_-_42129043 | 1.79 |
ENSMUST00000018965.3
|
Avpi1
|
arginine vasopressin-induced 1 |
| chr16_+_45158725 | 1.77 |
ENSMUST00000023343.3
|
Atg3
|
autophagy related 3 |
| chr11_-_96916448 | 1.70 |
ENSMUST00000103152.4
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr5_-_45639501 | 1.69 |
ENSMUST00000016023.7
|
Fam184b
|
family with sequence similarity 184, member B |
| chr8_+_27042555 | 1.66 |
ENSMUST00000033875.8
ENSMUST00000098851.4 |
Prosc
|
proline synthetase co-transcribed |
| chr1_+_5083105 | 1.65 |
ENSMUST00000044369.7
|
Atp6v1h
|
ATPase, H+ transporting, lysosomal V1 subunit H |
| chr13_-_92131494 | 1.64 |
ENSMUST00000099326.3
ENSMUST00000146492.1 |
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr11_-_88864534 | 1.63 |
ENSMUST00000018572.4
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr9_+_46012822 | 1.60 |
ENSMUST00000120463.2
ENSMUST00000120247.1 |
Sik3
|
SIK family kinase 3 |
| chr17_-_26939464 | 1.59 |
ENSMUST00000025027.8
ENSMUST00000114935.1 |
Cuta
|
cutA divalent cation tolerance homolog (E. coli) |
| chr16_-_45158650 | 1.57 |
ENSMUST00000023344.3
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr19_+_4855129 | 1.55 |
ENSMUST00000119694.1
|
Ctsf
|
cathepsin F |
| chr3_+_90052814 | 1.52 |
ENSMUST00000160640.1
ENSMUST00000029552.6 ENSMUST00000162114.1 ENSMUST00000068798.6 |
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr4_+_97772734 | 1.50 |
ENSMUST00000152023.1
|
Nfia
|
nuclear factor I/A |
| chr9_-_91365756 | 1.49 |
ENSMUST00000034927.6
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr2_-_130424242 | 1.48 |
ENSMUST00000089581.4
|
Pced1a
|
PC-esterase domain containing 1A |
| chr2_+_90847149 | 1.47 |
ENSMUST00000136872.1
|
Mtch2
|
mitochondrial carrier homolog 2 (C. elegans) |
| chr11_-_96916407 | 1.44 |
ENSMUST00000130774.1
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr3_-_107458895 | 1.44 |
ENSMUST00000009617.8
|
Kcnc4
|
potassium voltage gated channel, Shaw-related subfamily, member 4 |
| chr10_-_115251407 | 1.43 |
ENSMUST00000020339.8
|
Tbc1d15
|
TBC1 domain family, member 15 |
| chr6_+_13069758 | 1.41 |
ENSMUST00000124234.1
ENSMUST00000142211.1 ENSMUST00000031556.7 |
Tmem106b
|
transmembrane protein 106B |
| chr2_+_90847207 | 1.39 |
ENSMUST00000150232.1
ENSMUST00000111467.3 |
Mtch2
|
mitochondrial carrier homolog 2 (C. elegans) |
| chr11_-_50325599 | 1.38 |
ENSMUST00000179865.1
ENSMUST00000020637.8 |
Canx
|
calnexin |
| chr10_-_95416850 | 1.36 |
ENSMUST00000020215.9
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr10_-_95417099 | 1.35 |
ENSMUST00000135822.1
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr11_-_120617887 | 1.32 |
ENSMUST00000106188.3
ENSMUST00000026129.9 |
Pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr3_-_89089955 | 1.32 |
ENSMUST00000166687.1
|
Rusc1
|
RUN and SH3 domain containing 1 |
| chr8_-_106136792 | 1.28 |
ENSMUST00000146940.1
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr5_+_28165690 | 1.26 |
ENSMUST00000036177.7
|
En2
|
engrailed 2 |
| chr3_-_88296838 | 1.25 |
ENSMUST00000010682.3
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr2_-_37703275 | 1.25 |
ENSMUST00000072186.5
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr2_+_121449362 | 1.24 |
ENSMUST00000110615.1
ENSMUST00000099475.5 |
Serf2
|
small EDRK-rich factor 2 |
| chr8_-_106136890 | 1.23 |
ENSMUST00000115979.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr7_+_28766747 | 1.23 |
ENSMUST00000170068.1
ENSMUST00000072965.4 |
Sirt2
|
sirtuin 2 |
| chr17_-_65613521 | 1.23 |
ENSMUST00000024897.8
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr13_+_104109752 | 1.22 |
ENSMUST00000160322.1
ENSMUST00000159574.1 |
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr10_+_97693053 | 1.21 |
ENSMUST00000060703.4
|
Ccer1
|
coiled coil glutamate rich protein 1 |
| chr7_-_99182681 | 1.20 |
ENSMUST00000033001.4
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr9_+_122572493 | 1.19 |
ENSMUST00000181682.1
ENSMUST00000181107.1 ENSMUST00000181719.1 |
9530059O14Rik
|
RIKEN cDNA 9530059O14 gene |
| chr3_-_88296979 | 1.19 |
ENSMUST00000107556.3
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr7_-_28766469 | 1.18 |
ENSMUST00000085851.5
ENSMUST00000032815.4 |
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta |
| chr2_+_28641227 | 1.16 |
ENSMUST00000028155.5
ENSMUST00000113869.1 ENSMUST00000113867.2 |
Tsc1
|
tuberous sclerosis 1 |
| chr7_+_27195781 | 1.16 |
ENSMUST00000108379.1
ENSMUST00000179391.1 |
BC024978
|
cDNA sequence BC024978 |
| chr19_+_41981709 | 1.13 |
ENSMUST00000026170.1
|
Ubtd1
|
ubiquitin domain containing 1 |
| chr15_+_102921103 | 1.13 |
ENSMUST00000001700.6
|
Hoxc13
|
homeobox C13 |
| chr11_-_102296618 | 1.13 |
ENSMUST00000107132.2
ENSMUST00000073234.2 |
Atxn7l3
|
ataxin 7-like 3 |
| chr13_-_104109576 | 1.12 |
ENSMUST00000109315.3
|
Nln
|
neurolysin (metallopeptidase M3 family) |
| chr12_-_56535047 | 1.09 |
ENSMUST00000178477.2
|
Nkx2-1
|
NK2 homeobox 1 |
| chr16_-_5049882 | 1.09 |
ENSMUST00000023189.7
ENSMUST00000115844.1 |
Glyr1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr7_+_46796088 | 1.09 |
ENSMUST00000006774.4
ENSMUST00000165031.1 |
Gtf2h1
|
general transcription factor II H, polypeptide 1 |
| chr6_+_94500313 | 1.09 |
ENSMUST00000061118.8
|
Slc25a26
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
| chr13_+_104109737 | 1.08 |
ENSMUST00000044385.7
|
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr2_-_38926217 | 1.07 |
ENSMUST00000076275.4
ENSMUST00000142130.1 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr2_-_163419508 | 1.07 |
ENSMUST00000046908.3
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr16_-_97962581 | 1.06 |
ENSMUST00000113734.2
ENSMUST00000052089.7 ENSMUST00000063605.7 |
Zbtb21
|
zinc finger and BTB domain containing 21 |
| chr11_+_98937669 | 1.05 |
ENSMUST00000107475.2
ENSMUST00000068133.3 |
Rara
|
retinoic acid receptor, alpha |
| chr7_-_99141068 | 1.05 |
ENSMUST00000037968.8
|
Uvrag
|
UV radiation resistance associated gene |
| chr1_+_84839833 | 1.03 |
ENSMUST00000097672.3
|
Fbxo36
|
F-box protein 36 |
| chr6_-_52191695 | 1.03 |
ENSMUST00000101395.2
|
Hoxa4
|
homeobox A4 |
| chr10_+_121365078 | 1.03 |
ENSMUST00000040344.6
|
Gns
|
glucosamine (N-acetyl)-6-sulfatase |
| chr9_-_121857952 | 1.03 |
ENSMUST00000060251.6
|
Higd1a
|
HIG1 domain family, member 1A |
| chr7_-_28741780 | 1.03 |
ENSMUST00000056078.8
|
Mrps12
|
mitochondrial ribosomal protein S12 |
| chr7_-_27166732 | 1.02 |
ENSMUST00000080058.4
|
Egln2
|
EGL nine homolog 2 (C. elegans) |
| chr11_+_82388900 | 1.02 |
ENSMUST00000054245.4
ENSMUST00000092852.2 |
Tmem132e
|
transmembrane protein 132E |
| chr7_+_44850393 | 1.01 |
ENSMUST00000136232.1
|
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr16_-_45158566 | 0.99 |
ENSMUST00000181177.1
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr3_+_94954075 | 0.98 |
ENSMUST00000107260.2
ENSMUST00000142311.1 ENSMUST00000137088.1 ENSMUST00000152869.1 ENSMUST00000107254.1 ENSMUST00000107253.1 |
Rfx5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr6_+_29768443 | 0.98 |
ENSMUST00000166718.1
ENSMUST00000102995.2 ENSMUST00000115242.2 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr4_+_118428078 | 0.97 |
ENSMUST00000006557.6
ENSMUST00000167636.1 ENSMUST00000102673.4 |
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr5_+_76951307 | 0.97 |
ENSMUST00000031160.9
ENSMUST00000120912.1 ENSMUST00000117536.1 |
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr7_-_27166413 | 0.96 |
ENSMUST00000108382.1
|
Egln2
|
EGL nine homolog 2 (C. elegans) |
| chr5_+_76951382 | 0.96 |
ENSMUST00000141687.1
|
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr7_+_141455198 | 0.96 |
ENSMUST00000164016.1
ENSMUST00000064151.6 ENSMUST00000169665.1 |
Pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr7_-_28741749 | 0.95 |
ENSMUST00000171183.1
|
Mrps12
|
mitochondrial ribosomal protein S12 |
| chr1_-_183297008 | 0.95 |
ENSMUST00000057062.5
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr17_-_56626872 | 0.94 |
ENSMUST00000047226.8
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chr6_+_108660616 | 0.94 |
ENSMUST00000032194.4
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr17_+_45563928 | 0.94 |
ENSMUST00000041353.6
|
Slc35b2
|
solute carrier family 35, member B2 |
| chr11_-_118569910 | 0.93 |
ENSMUST00000136551.1
|
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr10_-_56228636 | 0.93 |
ENSMUST00000099739.3
|
Tbc1d32
|
TBC1 domain family, member 32 |
| chr16_-_45158624 | 0.93 |
ENSMUST00000180636.1
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr10_-_62486772 | 0.92 |
ENSMUST00000105447.3
|
Vps26a
|
vacuolar protein sorting 26 homolog A (yeast) |
| chr5_-_116591811 | 0.92 |
ENSMUST00000076124.5
|
Srrm4
|
serine/arginine repetitive matrix 4 |
| chr17_+_74528279 | 0.91 |
ENSMUST00000180037.1
ENSMUST00000024879.6 |
Birc6
|
baculoviral IAP repeat-containing 6 |
| chr11_-_98438941 | 0.90 |
ENSMUST00000002655.7
|
Mien1
|
migration and invasion enhancer 1 |
| chr7_-_27396542 | 0.90 |
ENSMUST00000108363.1
|
Sptbn4
|
spectrin beta, non-erythrocytic 4 |
| chr11_+_70844745 | 0.90 |
ENSMUST00000076270.6
ENSMUST00000179114.1 ENSMUST00000100928.4 ENSMUST00000177731.1 ENSMUST00000108533.3 ENSMUST00000081362.6 ENSMUST00000178245.1 |
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr13_-_54687696 | 0.89 |
ENSMUST00000177950.1
ENSMUST00000146931.1 |
Rnf44
|
ring finger protein 44 |
| chr9_+_56937462 | 0.87 |
ENSMUST00000034827.8
|
Imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr11_-_17211504 | 0.85 |
ENSMUST00000020317.7
|
Pno1
|
partner of NOB1 homolog (S. cerevisiae) |
| chr1_+_75142775 | 0.85 |
ENSMUST00000097694.4
|
Fam134a
|
family with sequence similarity 134, member A |
| chr17_+_46383725 | 0.85 |
ENSMUST00000113481.1
ENSMUST00000138127.1 |
Zfp318
|
zinc finger protein 318 |
| chr16_+_30065333 | 0.84 |
ENSMUST00000023171.7
|
Hes1
|
hairy and enhancer of split 1 (Drosophila) |
| chrX_+_139610612 | 0.84 |
ENSMUST00000113026.1
|
Rnf128
|
ring finger protein 128 |
| chr10_+_67535465 | 0.83 |
ENSMUST00000145754.1
|
Egr2
|
early growth response 2 |
| chr6_-_125165707 | 0.83 |
ENSMUST00000118875.1
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr9_-_91365778 | 0.82 |
ENSMUST00000065360.3
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr19_-_60581013 | 0.81 |
ENSMUST00000111460.3
ENSMUST00000081790.7 ENSMUST00000166712.1 |
Cacul1
|
CDK2 associated, cullin domain 1 |
| chr9_-_75409951 | 0.81 |
ENSMUST00000049355.10
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr1_-_75180349 | 0.81 |
ENSMUST00000027396.8
|
Abcb6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr1_-_75219245 | 0.81 |
ENSMUST00000079464.6
|
Tuba4a
|
tubulin, alpha 4A |
| chr17_+_56326045 | 0.81 |
ENSMUST00000139679.1
ENSMUST00000025036.4 ENSMUST00000086835.5 |
Kdm4b
|
lysine (K)-specific demethylase 4B |
| chr19_+_38836561 | 0.80 |
ENSMUST00000037302.5
|
Tbc1d12
|
TBC1D12: TBC1 domain family, member 12 |
| chr9_+_75410145 | 0.80 |
ENSMUST00000180533.1
ENSMUST00000180574.1 |
4933433G15Rik
|
RIKEN cDNA 4933433G15 gene |
| chr9_+_58582397 | 0.78 |
ENSMUST00000176557.1
ENSMUST00000114121.4 ENSMUST00000177064.1 |
Nptn
|
neuroplastin |
| chr3_+_108186332 | 0.78 |
ENSMUST00000050909.6
ENSMUST00000106659.2 ENSMUST00000106656.1 ENSMUST00000106661.2 |
Amigo1
|
adhesion molecule with Ig like domain 1 |
| chr17_-_56609689 | 0.78 |
ENSMUST00000052832.5
|
2410015M20Rik
|
RIKEN cDNA 2410015M20 gene |
| chr11_+_97703394 | 0.76 |
ENSMUST00000103147.4
|
Psmb3
|
proteasome (prosome, macropain) subunit, beta type 3 |
| chr16_-_45158453 | 0.76 |
ENSMUST00000181750.1
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr16_-_23225334 | 0.72 |
ENSMUST00000055369.4
|
BC106179
|
cDNA sequence BC106179 |
| chr17_-_27820445 | 0.72 |
ENSMUST00000114859.1
|
D17Wsu92e
|
DNA segment, Chr 17, Wayne State University 92, expressed |
| chr7_-_126583177 | 0.71 |
ENSMUST00000098036.2
ENSMUST00000032962.4 |
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr17_+_34647128 | 0.71 |
ENSMUST00000015605.8
ENSMUST00000182587.1 |
Atf6b
|
activating transcription factor 6 beta |
| chr13_-_55321928 | 0.71 |
ENSMUST00000035242.7
|
Rab24
|
RAB24, member RAS oncogene family |
| chr1_-_17097839 | 0.70 |
ENSMUST00000038382.4
|
Jph1
|
junctophilin 1 |
| chr3_+_96727611 | 0.70 |
ENSMUST00000029740.9
|
Rnf115
|
ring finger protein 115 |
| chr3_-_94786430 | 0.70 |
ENSMUST00000107272.1
|
Cgn
|
cingulin |
| chr11_-_102697710 | 0.69 |
ENSMUST00000164506.2
ENSMUST00000092569.6 |
Ccdc43
|
coiled-coil domain containing 43 |
| chr14_+_3412614 | 0.68 |
ENSMUST00000170123.1
|
Gm10409
|
predicted gene 10409 |
| chr3_+_41564880 | 0.68 |
ENSMUST00000168086.1
|
Phf17
|
PHD finger protein 17 |
| chr16_-_45158183 | 0.67 |
ENSMUST00000114600.1
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr6_+_110645572 | 0.67 |
ENSMUST00000071076.6
ENSMUST00000172951.1 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr13_-_37994111 | 0.67 |
ENSMUST00000021864.6
|
Ssr1
|
signal sequence receptor, alpha |
| chr11_+_17211912 | 0.67 |
ENSMUST00000046955.6
|
Wdr92
|
WD repeat domain 92 |
| chr16_-_44139196 | 0.67 |
ENSMUST00000063661.6
ENSMUST00000114666.2 |
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr9_+_107569112 | 0.64 |
ENSMUST00000010191.7
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr11_+_101087277 | 0.64 |
ENSMUST00000107302.1
ENSMUST00000107303.3 ENSMUST00000017945.8 ENSMUST00000149597.1 |
Mlx
|
MAX-like protein X |
| chr4_-_108833608 | 0.62 |
ENSMUST00000102742.4
|
Btf3l4
|
basic transcription factor 3-like 4 |
| chr17_+_34032071 | 0.61 |
ENSMUST00000174299.1
ENSMUST00000173554.1 |
Rxrb
|
retinoid X receptor beta |
| chr19_+_4756557 | 0.61 |
ENSMUST00000036744.7
|
Rbm4b
|
RNA binding motif protein 4B |
| chr17_-_27820534 | 0.61 |
ENSMUST00000075076.4
ENSMUST00000114863.2 |
D17Wsu92e
|
DNA segment, Chr 17, Wayne State University 92, expressed |
| chr17_-_79896028 | 0.61 |
ENSMUST00000068282.5
ENSMUST00000112437.1 |
Atl2
|
atlastin GTPase 2 |
| chr17_+_34647187 | 0.59 |
ENSMUST00000173984.1
|
Atf6b
|
activating transcription factor 6 beta |
| chr15_+_98167806 | 0.59 |
ENSMUST00000031914.4
|
AI836003
|
expressed sequence AI836003 |
| chr19_+_41911851 | 0.59 |
ENSMUST00000011896.6
|
Pgam1
|
phosphoglycerate mutase 1 |
| chr19_-_41981119 | 0.59 |
ENSMUST00000026168.2
ENSMUST00000171561.1 |
Mms19
|
MMS19 (MET18 S. cerevisiae) |
| chr7_-_35802968 | 0.59 |
ENSMUST00000061586.4
|
Zfp507
|
zinc finger protein 507 |
| chr10_+_22158566 | 0.58 |
ENSMUST00000181645.1
ENSMUST00000105522.2 |
Raet1e
H60b
|
retinoic acid early transcript 1E histocompatibility 60b |
| chr11_+_120348678 | 0.57 |
ENSMUST00000143813.1
|
0610009L18Rik
|
RIKEN cDNA 0610009L18 gene |
| chr1_-_82291370 | 0.57 |
ENSMUST00000069799.2
|
Irs1
|
insulin receptor substrate 1 |
| chr2_+_146221921 | 0.56 |
ENSMUST00000089257.4
|
Insm1
|
insulinoma-associated 1 |
| chr6_-_39377681 | 0.55 |
ENSMUST00000090243.4
|
Slc37a3
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 3 |
| chr11_+_69991061 | 0.55 |
ENSMUST00000018711.8
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr11_-_52000432 | 0.54 |
ENSMUST00000020657.6
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr7_-_68749170 | 0.54 |
ENSMUST00000118110.1
ENSMUST00000048068.7 |
Arrdc4
|
arrestin domain containing 4 |
| chr13_+_55321991 | 0.54 |
ENSMUST00000021942.6
|
Prelid1
|
PRELI domain containing 1 |
| chr13_-_54687644 | 0.53 |
ENSMUST00000129881.1
|
Rnf44
|
ring finger protein 44 |
| chr9_-_57606234 | 0.53 |
ENSMUST00000045068.8
|
Cplx3
|
complexin 3 |
| chr8_+_69791163 | 0.52 |
ENSMUST00000034326.6
|
Atp13a1
|
ATPase type 13A1 |
| chr2_+_37452231 | 0.51 |
ENSMUST00000148470.1
ENSMUST00000066055.3 ENSMUST00000112920.1 |
Rabgap1
|
RAB GTPase activating protein 1 |
| chr17_+_74528467 | 0.51 |
ENSMUST00000182944.1
ENSMUST00000182597.1 ENSMUST00000182133.1 ENSMUST00000183224.1 |
Birc6
|
baculoviral IAP repeat-containing 6 |
| chr4_-_148038769 | 0.50 |
ENSMUST00000030879.5
ENSMUST00000137724.1 |
Clcn6
|
chloride channel 6 |
| chr12_-_85270564 | 0.49 |
ENSMUST00000019378.6
ENSMUST00000166821.1 |
Mlh3
|
mutL homolog 3 (E coli) |
| chr7_-_46795881 | 0.49 |
ENSMUST00000107653.1
ENSMUST00000107654.1 ENSMUST00000014562.7 ENSMUST00000152759.1 |
Hps5
|
Hermansky-Pudlak syndrome 5 homolog (human) |
| chr11_-_64436653 | 0.49 |
ENSMUST00000177999.1
|
F930015N05Rik
|
RIKEN cDNA F930015N05 gene |
| chr10_+_67535493 | 0.49 |
ENSMUST00000145936.1
|
Egr2
|
early growth response 2 |
| chr6_+_108660772 | 0.48 |
ENSMUST00000163617.1
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr11_+_60417238 | 0.48 |
ENSMUST00000070681.6
|
Gid4
|
GID complex subunit 4, VID24 homolog (S. cerevisiae) |
| chr6_-_6217023 | 0.48 |
ENSMUST00000015256.8
|
Slc25a13
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
| chr5_-_106458440 | 0.47 |
ENSMUST00000086795.6
|
Barhl2
|
BarH-like 2 (Drosophila) |
| chr16_-_4790220 | 0.45 |
ENSMUST00000118703.1
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr15_+_80255184 | 0.45 |
ENSMUST00000109605.3
|
Atf4
|
activating transcription factor 4 |
| chr1_+_23762003 | 0.45 |
ENSMUST00000140583.1
|
B3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr16_-_92466081 | 0.44 |
ENSMUST00000060005.8
|
Rcan1
|
regulator of calcineurin 1 |
| chr4_+_139352587 | 0.43 |
ENSMUST00000042096.7
ENSMUST00000179784.1 ENSMUST00000082262.7 ENSMUST00000147999.1 |
Emc1
Ubr4
|
ER membrane protein complex subunit 1 ubiquitin protein ligase E3 component n-recognin 4 |
| chr12_-_3309912 | 0.43 |
ENSMUST00000021001.8
|
Rab10
|
RAB10, member RAS oncogene family |
Network of associatons between targets according to the STRING database.
First level regulatory network of Arnt
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 1.3 | 4.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 1.1 | 3.3 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 1.1 | 18.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.6 | 1.8 | GO:0006059 | hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) fructose biosynthetic process(GO:0046370) |
| 0.6 | 2.9 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.6 | 13.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.5 | 3.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.5 | 1.4 | GO:1904057 | negative regulation of sensory perception of pain(GO:1904057) |
| 0.5 | 3.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.5 | 1.8 | GO:0015744 | succinate transport(GO:0015744) |
| 0.4 | 1.2 | GO:0006407 | rRNA export from nucleus(GO:0006407) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.4 | 1.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.4 | 1.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.4 | 1.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.4 | 1.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.3 | 3.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.3 | 1.3 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.3 | 1.3 | GO:0021594 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.3 | 1.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 | 3.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 2.9 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.2 | 1.9 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 1.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.2 | 0.6 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.2 | 1.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 1.9 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 1.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 3.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 0.9 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 1.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.2 | 0.8 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.3 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.2 | 1.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 2.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 1.2 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.1 | 1.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.7 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.4 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 | 0.6 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.7 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.9 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 0.4 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 1.9 | GO:0007567 | parturition(GO:0007567) ductus arteriosus closure(GO:0097070) |
| 0.1 | 0.6 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 1.8 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 1.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 4.5 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.1 | 0.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 4.8 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 0.6 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.7 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 2.7 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 1.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.4 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 1.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 1.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 2.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.3 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.1 | 0.3 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 1.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 1.6 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.1 | 0.3 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 1.0 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 1.4 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.1 | 2.0 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.1 | 0.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 1.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.6 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 0.4 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 1.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 0.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.4 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.6 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.4 | GO:1903751 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 2.3 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 0.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 1.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.8 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.3 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.7 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.9 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.8 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 2.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0045917 | positive regulation of complement activation(GO:0045917) progesterone receptor signaling pathway(GO:0050847) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.6 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.6 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 1.4 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 2.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.4 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 1.5 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 4.8 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 3.7 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.3 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 1.0 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.5 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 1.8 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.9 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.6 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.5 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.0 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.0 | 0.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 17.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.1 | 3.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.6 | 2.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.4 | 1.6 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.4 | 1.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.3 | 0.9 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 0.7 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 0.9 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.2 | 1.6 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.2 | 1.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 1.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 2.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.1 | 1.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.6 | GO:0071817 | MMXD complex(GO:0071817) CIA complex(GO:0097361) |
| 0.1 | 1.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.5 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.3 | GO:0030934 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 0.1 | 0.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 2.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 3.0 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.6 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 2.0 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 4.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.8 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 3.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 11.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 3.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 2.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 7.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 4.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 2.3 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.2 | GO:0043197 | dendritic spine(GO:0043197) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.6 | 1.8 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.6 | 1.8 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.5 | 2.7 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.5 | 3.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.5 | 1.9 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.4 | 1.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.4 | 1.2 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.4 | 2.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 1.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 0.9 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.3 | 1.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.3 | 2.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 0.9 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.3 | 1.9 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.3 | 3.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 0.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 1.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 0.7 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 0.6 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.2 | 1.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 4.9 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.2 | 0.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 2.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 0.6 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 1.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 1.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 1.9 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 0.5 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 1.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 1.3 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 2.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 1.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 2.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 3.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 3.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 1.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 1.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 16.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 2.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.4 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.7 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.8 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.9 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 1.1 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 2.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.0 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 2.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 2.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 3.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 6.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 3.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 3.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 3.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 5.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 2.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 4.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 3.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 4.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.9 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 1.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |