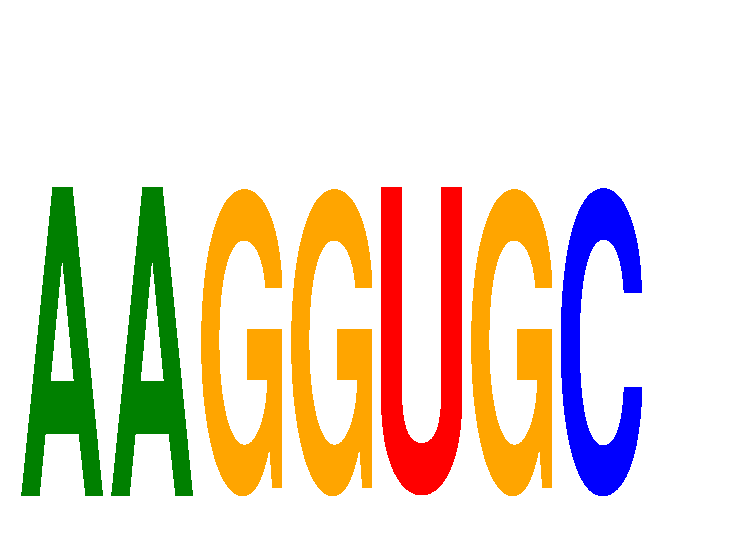Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for AAGGUGC
Z-value: 0.33
miRNA associated with seed AAGGUGC
| Name | miRBASE accession |
|---|---|
|
mmu-miR-18a-5p
|
MIMAT0000528 |
|
mmu-miR-18b-5p
|
MIMAT0004858 |
Activity profile of AAGGUGC motif
Sorted Z-values of AAGGUGC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_76669811 | 2.07 |
ENSMUST00000037824.4
|
Foxh1
|
forkhead box H1 |
| chr16_-_92826004 | 1.00 |
ENSMUST00000023673.7
|
Runx1
|
runt related transcription factor 1 |
| chr16_-_22163299 | 0.92 |
ENSMUST00000100052.4
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr7_-_116038734 | 0.84 |
ENSMUST00000166877.1
|
Sox6
|
SRY-box containing gene 6 |
| chr5_-_73256555 | 0.74 |
ENSMUST00000171179.1
ENSMUST00000101127.5 |
Fryl
Fryl
|
furry homolog-like (Drosophila) furry homolog-like (Drosophila) |
| chr10_-_78244602 | 0.74 |
ENSMUST00000000384.6
|
Trappc10
|
trafficking protein particle complex 10 |
| chr1_-_161876656 | 0.71 |
ENSMUST00000048377.5
|
Suco
|
SUN domain containing ossification factor |
| chr2_+_118111876 | 0.69 |
ENSMUST00000039559.8
|
Thbs1
|
thrombospondin 1 |
| chrX_+_9199865 | 0.68 |
ENSMUST00000069763.2
|
Lancl3
|
LanC lantibiotic synthetase component C-like 3 (bacterial) |
| chr2_-_34913976 | 0.67 |
ENSMUST00000028232.3
|
Phf19
|
PHD finger protein 19 |
| chr17_-_29888570 | 0.57 |
ENSMUST00000171691.1
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr2_+_156420837 | 0.54 |
ENSMUST00000103137.3
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1 |
| chr2_+_164074122 | 0.53 |
ENSMUST00000018353.7
|
Stk4
|
serine/threonine kinase 4 |
| chr1_+_170644523 | 0.53 |
ENSMUST00000046792.8
|
Olfml2b
|
olfactomedin-like 2B |
| chr4_-_134018829 | 0.51 |
ENSMUST00000051674.2
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr1_-_97661950 | 0.49 |
ENSMUST00000053033.7
ENSMUST00000149927.1 |
D1Ertd622e
|
DNA segment, Chr 1, ERATO Doi 622, expressed |
| chr3_+_105452326 | 0.44 |
ENSMUST00000098761.3
|
Kcnd3
|
potassium voltage-gated channel, Shal-related family, member 3 |
| chr8_-_35495487 | 0.41 |
ENSMUST00000033927.6
|
Eri1
|
exoribonuclease 1 |
| chr5_+_96997676 | 0.41 |
ENSMUST00000112974.1
ENSMUST00000035635.7 |
Bmp2k
|
BMP2 inducible kinase |
| chr5_+_75574916 | 0.40 |
ENSMUST00000144270.1
ENSMUST00000005815.6 |
Kit
|
kit oncogene |
| chr8_-_120589304 | 0.38 |
ENSMUST00000034278.5
|
Gins2
|
GINS complex subunit 2 (Psf2 homolog) |
| chr14_+_47472547 | 0.35 |
ENSMUST00000168833.1
ENSMUST00000163324.1 ENSMUST00000043112.7 |
Fbxo34
|
F-box protein 34 |
| chr19_+_53140430 | 0.34 |
ENSMUST00000111741.2
|
Add3
|
adducin 3 (gamma) |
| chr15_+_80711292 | 0.34 |
ENSMUST00000067689.7
|
Tnrc6b
|
trinucleotide repeat containing 6b |
| chr15_+_73723131 | 0.34 |
ENSMUST00000165541.1
ENSMUST00000167582.1 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr6_+_119236507 | 0.33 |
ENSMUST00000037434.6
|
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr13_+_109903089 | 0.33 |
ENSMUST00000120664.1
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr10_+_40349265 | 0.32 |
ENSMUST00000044672.4
ENSMUST00000095743.2 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr1_-_63114516 | 0.27 |
ENSMUST00000097718.2
|
Ino80d
|
INO80 complex subunit D |
| chr10_-_53379816 | 0.26 |
ENSMUST00000095691.5
|
Cep85l
|
centrosomal protein 85-like |
| chr9_-_14381242 | 0.25 |
ENSMUST00000167549.1
|
Endod1
|
endonuclease domain containing 1 |
| chr11_+_29373618 | 0.24 |
ENSMUST00000040182.6
ENSMUST00000109477.1 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr15_-_12592556 | 0.24 |
ENSMUST00000075317.5
|
Pdzd2
|
PDZ domain containing 2 |
| chr10_-_19015347 | 0.22 |
ENSMUST00000019997.4
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr16_-_78376758 | 0.21 |
ENSMUST00000023570.7
|
Btg3
|
B cell translocation gene 3 |
| chr3_-_9610074 | 0.21 |
ENSMUST00000041124.7
|
Zfp704
|
zinc finger protein 704 |
| chr3_-_30969399 | 0.21 |
ENSMUST00000177992.1
ENSMUST00000129817.2 ENSMUST00000168645.1 ENSMUST00000108255.1 ENSMUST00000064718.5 ENSMUST00000099163.3 |
Phc3
|
polyhomeotic-like 3 (Drosophila) |
| chr6_+_52713729 | 0.21 |
ENSMUST00000080723.4
ENSMUST00000149588.1 |
Tax1bp1
|
Tax1 (human T cell leukemia virus type I) binding protein 1 |
| chr1_+_74409376 | 0.21 |
ENSMUST00000027366.6
|
Vil1
|
villin 1 |
| chr12_+_52516077 | 0.20 |
ENSMUST00000110725.1
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr12_-_15816762 | 0.20 |
ENSMUST00000020922.7
|
Trib2
|
tribbles homolog 2 (Drosophila) |
| chr6_-_25690729 | 0.20 |
ENSMUST00000054867.6
|
Gpr37
|
G protein-coupled receptor 37 |
| chr9_+_121760000 | 0.19 |
ENSMUST00000093772.3
|
Zfp651
|
zinc finger protein 651 |
| chr3_+_60501252 | 0.19 |
ENSMUST00000099087.2
|
Mbnl1
|
muscleblind-like 1 (Drosophila) |
| chr19_-_59943647 | 0.19 |
ENSMUST00000171986.1
|
Rab11fip2
|
RAB11 family interacting protein 2 (class I) |
| chr18_+_53472981 | 0.18 |
ENSMUST00000115399.1
|
Prdm6
|
PR domain containing 6 |
| chr5_+_137518880 | 0.18 |
ENSMUST00000031727.7
|
Gigyf1
|
GRB10 interacting GYF protein 1 |
| chr13_-_111808938 | 0.17 |
ENSMUST00000109267.2
|
Map3k1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr5_-_69592311 | 0.16 |
ENSMUST00000031117.6
|
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr13_+_60211933 | 0.16 |
ENSMUST00000060705.2
|
Gm5084
|
predicted gene 5084 |
| chr8_-_84687839 | 0.15 |
ENSMUST00000001975.4
|
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr13_+_41606216 | 0.15 |
ENSMUST00000129449.1
|
Tmem170b
|
transmembrane protein 170B |
| chr3_-_59344256 | 0.14 |
ENSMUST00000039419.6
|
Igsf10
|
immunoglobulin superfamily, member 10 |
| chr16_+_94085226 | 0.14 |
ENSMUST00000072182.7
|
Sim2
|
single-minded homolog 2 (Drosophila) |
| chr5_+_23434435 | 0.13 |
ENSMUST00000094962.2
ENSMUST00000115128.1 |
Kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr13_-_93499803 | 0.13 |
ENSMUST00000065537.7
|
Jmy
|
junction-mediating and regulatory protein |
| chrX_-_108834303 | 0.13 |
ENSMUST00000101283.3
ENSMUST00000150434.1 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr4_-_119492563 | 0.12 |
ENSMUST00000049994.7
|
Rimkla
|
ribosomal modification protein rimK-like family member A |
| chr13_-_41487306 | 0.12 |
ENSMUST00000021794.6
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9 |
| chr10_+_13552894 | 0.12 |
ENSMUST00000019944.8
|
Adat2
|
adenosine deaminase, tRNA-specific 2 |
| chr1_-_170215380 | 0.12 |
ENSMUST00000027979.7
ENSMUST00000123399.1 |
Uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr11_+_77686155 | 0.12 |
ENSMUST00000100802.4
ENSMUST00000181023.1 |
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr2_+_65845767 | 0.11 |
ENSMUST00000122912.1
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr11_+_32455362 | 0.10 |
ENSMUST00000051053.4
|
Ubtd2
|
ubiquitin domain containing 2 |
| chr9_-_53536728 | 0.10 |
ENSMUST00000118282.1
|
Atm
|
ataxia telangiectasia mutated homolog (human) |
| chr13_+_97241096 | 0.10 |
ENSMUST00000041623.7
|
Enc1
|
ectodermal-neural cortex 1 |
| chr2_+_19445632 | 0.08 |
ENSMUST00000028068.2
|
Ptf1a
|
pancreas specific transcription factor, 1a |
| chr7_-_126502380 | 0.08 |
ENSMUST00000167759.1
|
Atxn2l
|
ataxin 2-like |
| chr16_+_44139821 | 0.08 |
ENSMUST00000159514.1
ENSMUST00000161326.1 ENSMUST00000063520.8 ENSMUST00000063542.7 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr2_+_72285637 | 0.07 |
ENSMUST00000090824.5
ENSMUST00000135469.1 |
Zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr1_-_166002591 | 0.07 |
ENSMUST00000111429.4
ENSMUST00000176800.1 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr11_-_116086929 | 0.07 |
ENSMUST00000074628.6
ENSMUST00000106444.3 |
Wbp2
|
WW domain binding protein 2 |
| chr1_+_134560190 | 0.06 |
ENSMUST00000112198.1
ENSMUST00000112197.1 |
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chr12_+_3426857 | 0.05 |
ENSMUST00000111215.3
ENSMUST00000092003.5 ENSMUST00000144247.2 ENSMUST00000153102.2 |
Asxl2
|
additional sex combs like 2 (Drosophila) |
| chr10_+_80494835 | 0.05 |
ENSMUST00000051773.8
|
Onecut3
|
one cut domain, family member 3 |
| chr3_+_96697076 | 0.04 |
ENSMUST00000162778.2
ENSMUST00000064900.9 |
Pias3
|
protein inhibitor of activated STAT 3 |
| chr13_-_64153194 | 0.04 |
ENSMUST00000059817.4
ENSMUST00000117241.1 |
Zfp367
|
zinc finger protein 367 |
| chr3_-_27896360 | 0.04 |
ENSMUST00000058077.3
|
Tmem212
|
transmembrane protein 212 |
| chr1_-_161131428 | 0.03 |
ENSMUST00000111611.1
|
Klhl20
|
kelch-like 20 |
| chr12_+_84875769 | 0.03 |
ENSMUST00000171853.1
|
D030025P21Rik
|
RIKEN cDNA D030025P21 gene |
| chr11_-_100738153 | 0.03 |
ENSMUST00000155500.1
ENSMUST00000107364.1 ENSMUST00000019317.5 |
Rab5c
|
RAB5C, member RAS oncogene family |
| chr13_-_48870885 | 0.02 |
ENSMUST00000035540.7
|
Phf2
|
PHD finger protein 2 |
| chr16_-_52454074 | 0.01 |
ENSMUST00000023312.7
|
Alcam
|
activated leukocyte cell adhesion molecule |
| chr12_-_91590009 | 0.01 |
ENSMUST00000021345.6
|
Gtf2a1
|
general transcription factor II A, 1 |
| chr12_+_12262139 | 0.01 |
ENSMUST00000069066.6
ENSMUST00000069005.8 |
Fam49a
|
family with sequence similarity 49, member A |
| chr3_+_88142328 | 0.01 |
ENSMUST00000001455.6
ENSMUST00000119251.1 |
Mef2d
|
myocyte enhancer factor 2D |
| chr1_-_135585314 | 0.00 |
ENSMUST00000040599.8
ENSMUST00000067414.6 |
Nav1
|
neuron navigator 1 |
| chr8_-_94601720 | 0.00 |
ENSMUST00000034226.6
|
Fam192a
|
family with sequence similarity 192, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGGUGC
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.3 | 1.0 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.2 | 0.7 | GO:0010752 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of cGMP-mediated signaling(GO:0010752) |
| 0.1 | 0.5 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.7 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.4 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.1 | 0.6 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.8 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.2 | GO:0002631 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 0.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.5 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.4 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.4 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.1 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.2 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.6 | GO:0010714 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0035938 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 1.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |