Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
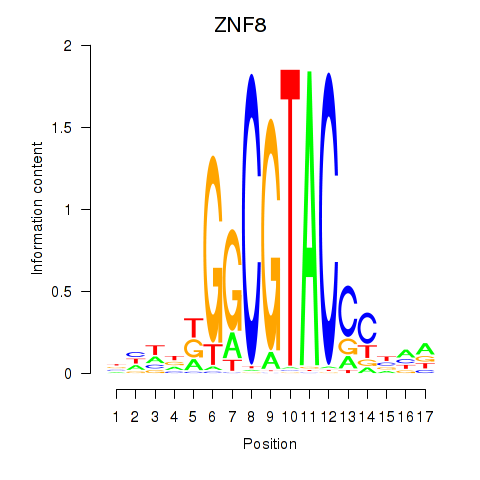
Results for ZNF8
Z-value: 0.55
Transcription factors associated with ZNF8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF8
|
ENSG00000083842.8 | zinc finger protein 8 |
|
ZNF8
|
ENSG00000273439.1 | zinc finger protein 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF8 | hg19_v2_chr19_+_58790314_58790358 | -0.42 | 4.1e-01 | Click! |
Activity profile of ZNF8 motif
Sorted Z-values of ZNF8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_79258444 | 0.42 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr12_+_79258547 | 0.35 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr5_-_43515125 | 0.28 |
ENST00000509489.1
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chr6_+_29691056 | 0.26 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr6_+_29691198 | 0.25 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr1_+_186344945 | 0.23 |
ENST00000419367.3
ENST00000287859.6 |
C1orf27
|
chromosome 1 open reading frame 27 |
| chr17_+_48585794 | 0.23 |
ENST00000576179.1
ENST00000419930.1 |
MYCBPAP
|
MYCBP associated protein |
| chr11_+_13690249 | 0.22 |
ENST00000532701.1
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr1_-_70671216 | 0.21 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr1_+_186344883 | 0.20 |
ENST00000367470.3
|
C1orf27
|
chromosome 1 open reading frame 27 |
| chr8_-_109260897 | 0.20 |
ENST00000521297.1
ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E
|
eukaryotic translation initiation factor 3, subunit E |
| chr9_-_32552551 | 0.20 |
ENST00000360538.2
ENST00000379858.1 |
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase |
| chr11_+_13690200 | 0.19 |
ENST00000354817.3
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr13_+_45563721 | 0.19 |
ENST00000361121.2
|
GPALPP1
|
GPALPP motifs containing 1 |
| chr4_-_52904425 | 0.19 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr1_+_70671363 | 0.18 |
ENST00000370951.1
ENST00000370950.3 ENST00000405432.1 ENST00000454435.2 |
SRSF11
|
serine/arginine-rich splicing factor 11 |
| chr12_-_46662888 | 0.18 |
ENST00000546893.1
|
SLC38A1
|
solute carrier family 38, member 1 |
| chr4_-_77069573 | 0.17 |
ENST00000264883.3
|
NUP54
|
nucleoporin 54kDa |
| chr13_+_53029564 | 0.16 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr12_-_46662772 | 0.15 |
ENST00000549049.1
ENST00000439706.1 ENST00000398637.5 |
SLC38A1
|
solute carrier family 38, member 1 |
| chr5_+_102455968 | 0.15 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr5_+_138677515 | 0.14 |
ENST00000265192.4
ENST00000511706.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr2_+_172290707 | 0.14 |
ENST00000375255.3
ENST00000539783.1 |
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr8_-_61429315 | 0.13 |
ENST00000530725.1
ENST00000532232.1 |
RP11-163N6.2
|
RP11-163N6.2 |
| chrX_+_102631248 | 0.13 |
ENST00000361298.4
ENST00000372645.3 ENST00000372635.1 |
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr11_+_46402583 | 0.12 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr10_+_135192695 | 0.12 |
ENST00000368539.4
ENST00000278060.5 ENST00000357296.3 |
PAOX
|
polyamine oxidase (exo-N4-amino) |
| chr17_+_48585902 | 0.12 |
ENST00000452039.1
|
MYCBPAP
|
MYCBP associated protein |
| chr16_+_68573116 | 0.12 |
ENST00000570495.1
ENST00000563169.2 ENST00000564323.1 ENST00000562156.1 ENST00000573685.1 |
ZFP90
|
ZFP90 zinc finger protein |
| chr10_-_1034237 | 0.12 |
ENST00000381466.1
|
AL359878.1
|
Uncharacterized protein |
| chr1_-_100715372 | 0.12 |
ENST00000370131.3
ENST00000370132.4 |
DBT
|
dihydrolipoamide branched chain transacylase E2 |
| chr13_+_53226963 | 0.12 |
ENST00000343788.6
ENST00000535397.1 ENST00000310528.8 |
SUGT1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr13_-_21750659 | 0.11 |
ENST00000400018.3
ENST00000314759.5 |
SKA3
|
spindle and kinetochore associated complex subunit 3 |
| chr2_-_172290482 | 0.11 |
ENST00000442541.1
ENST00000392599.2 ENST00000375258.4 |
METTL8
|
methyltransferase like 8 |
| chr12_-_74686314 | 0.11 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr6_+_88117683 | 0.10 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr1_-_235292250 | 0.10 |
ENST00000366607.4
|
TOMM20
|
translocase of outer mitochondrial membrane 20 homolog (yeast) |
| chr4_+_170541678 | 0.10 |
ENST00000360642.3
ENST00000512813.1 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr8_+_38244638 | 0.10 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr4_+_170541835 | 0.09 |
ENST00000504131.2
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr5_-_137911049 | 0.09 |
ENST00000297185.3
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin) |
| chr15_+_85523671 | 0.09 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr2_+_198365122 | 0.09 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr8_+_61429416 | 0.08 |
ENST00000262646.7
ENST00000531289.1 |
RAB2A
|
RAB2A, member RAS oncogene family |
| chr16_-_53537105 | 0.08 |
ENST00000568596.1
ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP
|
AKT interacting protein |
| chr10_+_1034338 | 0.07 |
ENST00000360803.4
ENST00000538293.1 |
GTPBP4
|
GTP binding protein 4 |
| chr1_+_25664408 | 0.07 |
ENST00000374358.4
|
TMEM50A
|
transmembrane protein 50A |
| chr13_-_25861530 | 0.07 |
ENST00000540661.1
|
MTMR6
|
myotubularin related protein 6 |
| chr2_+_169926047 | 0.07 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr4_-_186347099 | 0.06 |
ENST00000505357.1
ENST00000264689.6 |
UFSP2
|
UFM1-specific peptidase 2 |
| chr14_+_51706886 | 0.06 |
ENST00000457354.2
|
TMX1
|
thioredoxin-related transmembrane protein 1 |
| chr16_+_21964662 | 0.06 |
ENST00000561553.1
ENST00000565331.1 |
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II |
| chr4_+_170541660 | 0.06 |
ENST00000513761.1
ENST00000347613.4 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr5_-_93447333 | 0.06 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr10_+_1034646 | 0.05 |
ENST00000360059.5
ENST00000545048.1 |
GTPBP4
|
GTP binding protein 4 |
| chr17_+_48585745 | 0.05 |
ENST00000323776.5
|
MYCBPAP
|
MYCBP associated protein |
| chr1_+_211500129 | 0.05 |
ENST00000427925.2
ENST00000261464.5 |
TRAF5
|
TNF receptor-associated factor 5 |
| chr1_-_246670519 | 0.04 |
ENST00000388985.4
ENST00000490107.1 |
SMYD3
|
SET and MYND domain containing 3 |
| chr2_+_173600565 | 0.04 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chrX_+_128913906 | 0.03 |
ENST00000356892.3
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr2_+_173600514 | 0.03 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_240323439 | 0.03 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr3_-_46000146 | 0.03 |
ENST00000438446.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr22_-_30162924 | 0.03 |
ENST00000344318.3
ENST00000397781.3 |
ZMAT5
|
zinc finger, matrin-type 5 |
| chr6_+_116601265 | 0.03 |
ENST00000452085.3
|
DSE
|
dermatan sulfate epimerase |
| chr1_+_6615241 | 0.03 |
ENST00000333172.6
ENST00000328191.4 ENST00000351136.3 |
TAS1R1
|
taste receptor, type 1, member 1 |
| chr12_-_122750957 | 0.03 |
ENST00000451053.2
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr20_+_37377085 | 0.02 |
ENST00000243903.4
|
ACTR5
|
ARP5 actin-related protein 5 homolog (yeast) |
| chrX_+_1387693 | 0.02 |
ENST00000381529.3
ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr6_+_106959718 | 0.02 |
ENST00000369066.3
|
AIM1
|
absent in melanoma 1 |
| chr2_+_87144738 | 0.02 |
ENST00000559485.1
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr5_+_89854595 | 0.02 |
ENST00000405460.2
|
GPR98
|
G protein-coupled receptor 98 |
| chr7_+_1748798 | 0.01 |
ENST00000561626.1
|
ELFN1
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr10_-_13043697 | 0.01 |
ENST00000378825.3
|
CCDC3
|
coiled-coil domain containing 3 |
| chr11_-_58345569 | 0.01 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr19_+_52074502 | 0.01 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr11_+_46402482 | 0.01 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.4 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.5 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.2 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.5 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.3 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 0.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |