Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
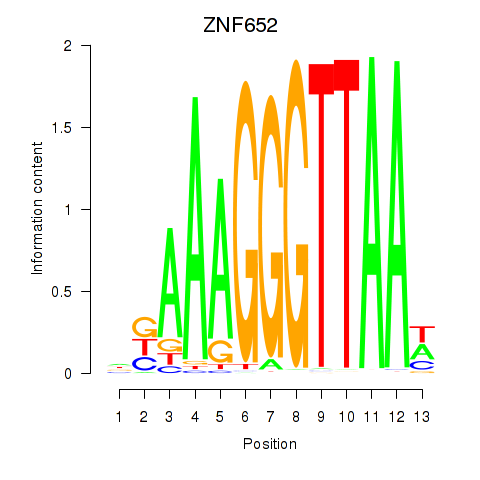
Results for ZNF652
Z-value: 1.00
Transcription factors associated with ZNF652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF652
|
ENSG00000198740.4 | zinc finger protein 652 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF652 | hg19_v2_chr17_-_47439437_47439533 | 0.89 | 1.8e-02 | Click! |
Activity profile of ZNF652 motif
Sorted Z-values of ZNF652 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_6662919 | 0.73 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr11_-_86383461 | 0.72 |
ENST00000532471.1
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr4_-_90757364 | 0.71 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr5_-_78281775 | 0.69 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr14_+_59104741 | 0.51 |
ENST00000395153.3
ENST00000335867.4 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr15_+_75640068 | 0.51 |
ENST00000565051.1
ENST00000564257.1 ENST00000567005.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr12_-_120241187 | 0.50 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr2_+_66918558 | 0.47 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr2_+_138722028 | 0.43 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr10_+_104263743 | 0.38 |
ENST00000369902.3
ENST00000369899.2 ENST00000423559.2 |
SUFU
|
suppressor of fused homolog (Drosophila) |
| chr12_-_49463620 | 0.37 |
ENST00000550675.1
|
RHEBL1
|
Ras homolog enriched in brain like 1 |
| chr4_-_90756769 | 0.36 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr2_-_203735484 | 0.36 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr12_+_6982725 | 0.35 |
ENST00000433346.1
|
LRRC23
|
leucine rich repeat containing 23 |
| chr2_-_36779411 | 0.34 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr12_-_6982442 | 0.33 |
ENST00000523102.1
ENST00000524270.1 ENST00000519357.1 |
SPSB2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr1_+_39670423 | 0.33 |
ENST00000536367.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr9_+_132096166 | 0.33 |
ENST00000436710.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr16_+_31483451 | 0.33 |
ENST00000565360.1
ENST00000361773.3 |
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr5_+_149865838 | 0.32 |
ENST00000519157.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr16_-_79634595 | 0.32 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr11_+_72975524 | 0.32 |
ENST00000540342.1
ENST00000542092.1 |
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr14_-_69262947 | 0.32 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr3_-_64431146 | 0.31 |
ENST00000569824.1
ENST00000485770.1 |
PRICKLE2
RP11-14D22.2
|
prickle homolog 2 (Drosophila) RP11-14D22.2 |
| chr17_-_4689727 | 0.31 |
ENST00000328739.5
ENST00000354194.4 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr19_+_53700340 | 0.31 |
ENST00000597550.1
ENST00000601072.1 |
CTD-2245F17.3
|
CTD-2245F17.3 |
| chr14_+_65182395 | 0.31 |
ENST00000554088.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr2_-_61244550 | 0.30 |
ENST00000421319.1
|
PUS10
|
pseudouridylate synthase 10 |
| chr1_-_8483723 | 0.30 |
ENST00000476556.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr19_+_10531150 | 0.30 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr1_+_145293371 | 0.29 |
ENST00000342960.5
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr1_+_39670360 | 0.29 |
ENST00000494012.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr2_-_73460334 | 0.28 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr10_-_105437909 | 0.28 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr11_+_72975578 | 0.27 |
ENST00000393592.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr11_-_89224638 | 0.27 |
ENST00000535633.1
ENST00000263317.4 |
NOX4
|
NADPH oxidase 4 |
| chr16_-_75284758 | 0.27 |
ENST00000561970.1
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr11_+_19798964 | 0.26 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr19_+_17326191 | 0.25 |
ENST00000595101.1
ENST00000596136.1 ENST00000379776.4 |
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr3_-_9994021 | 0.25 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr3_+_111393501 | 0.25 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr12_-_57443886 | 0.25 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr7_-_92465868 | 0.25 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr14_-_69262789 | 0.25 |
ENST00000557022.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr11_-_86383370 | 0.25 |
ENST00000526834.1
ENST00000359636.2 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr22_-_20255212 | 0.25 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr16_+_31483374 | 0.25 |
ENST00000394863.3
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr8_-_145018080 | 0.24 |
ENST00000354589.3
|
PLEC
|
plectin |
| chr19_-_37329254 | 0.24 |
ENST00000356725.4
|
ZNF790
|
zinc finger protein 790 |
| chr11_-_71823715 | 0.24 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr5_-_157002749 | 0.24 |
ENST00000517905.1
ENST00000430702.2 ENST00000394020.1 |
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr2_-_218808771 | 0.23 |
ENST00000449814.1
ENST00000171887.4 |
TNS1
|
tensin 1 |
| chr19_+_37998031 | 0.23 |
ENST00000586138.1
ENST00000588578.1 ENST00000587986.1 |
ZNF793
|
zinc finger protein 793 |
| chr15_+_75639951 | 0.23 |
ENST00000564784.1
ENST00000569035.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr17_+_38337491 | 0.23 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr16_-_18908196 | 0.23 |
ENST00000565324.1
ENST00000561947.1 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr14_+_85996471 | 0.23 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr11_-_118966167 | 0.22 |
ENST00000530167.1
|
H2AFX
|
H2A histone family, member X |
| chr11_+_72975559 | 0.22 |
ENST00000349767.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr3_+_193853927 | 0.22 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr10_-_28623368 | 0.22 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr8_-_18711866 | 0.22 |
ENST00000519851.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr4_-_77134742 | 0.22 |
ENST00000452464.2
|
SCARB2
|
scavenger receptor class B, member 2 |
| chr1_-_150780757 | 0.21 |
ENST00000271651.3
|
CTSK
|
cathepsin K |
| chr16_+_31470143 | 0.21 |
ENST00000457010.2
ENST00000563544.1 |
ARMC5
|
armadillo repeat containing 5 |
| chr11_+_107461948 | 0.21 |
ENST00000265840.7
ENST00000443271.2 |
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr16_-_71610985 | 0.21 |
ENST00000355962.4
|
TAT
|
tyrosine aminotransferase |
| chr15_-_34880646 | 0.21 |
ENST00000543376.1
|
GOLGA8A
|
golgin A8 family, member A |
| chr5_-_57854070 | 0.21 |
ENST00000504333.1
|
CTD-2117L12.1
|
Uncharacterized protein |
| chr9_+_130860583 | 0.21 |
ENST00000373064.5
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr5_+_180467259 | 0.21 |
ENST00000515271.1
|
BTNL9
|
butyrophilin-like 9 |
| chr3_+_119814070 | 0.20 |
ENST00000469070.1
|
RP11-18H7.1
|
RP11-18H7.1 |
| chr15_+_22368478 | 0.20 |
ENST00000332663.2
|
OR4M2
|
olfactory receptor, family 4, subfamily M, member 2 |
| chr11_-_130298888 | 0.20 |
ENST00000257359.6
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr19_-_3985455 | 0.20 |
ENST00000309311.6
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr16_+_31470179 | 0.20 |
ENST00000538189.1
ENST00000268314.4 |
ARMC5
|
armadillo repeat containing 5 |
| chr16_+_67927147 | 0.20 |
ENST00000291041.5
|
PSKH1
|
protein serine kinase H1 |
| chr22_-_37880543 | 0.20 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chrX_+_134887233 | 0.20 |
ENST00000443882.1
|
CT45A3
|
cancer/testis antigen family 45, member A3 |
| chr2_+_11052054 | 0.20 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr9_+_130860810 | 0.20 |
ENST00000433501.1
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr12_+_93964746 | 0.20 |
ENST00000536696.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr11_-_71823796 | 0.20 |
ENST00000545680.1
ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr8_+_77593474 | 0.20 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr14_+_22471345 | 0.20 |
ENST00000390446.3
|
TRAV18
|
T cell receptor alpha variable 18 |
| chr10_-_101190202 | 0.19 |
ENST00000543866.1
ENST00000370508.5 |
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chrX_+_48916497 | 0.19 |
ENST00000496529.2
ENST00000376396.3 ENST00000422185.2 ENST00000603986.1 ENST00000536628.2 |
CCDC120
|
coiled-coil domain containing 120 |
| chr8_-_125577940 | 0.19 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr17_-_19290117 | 0.19 |
ENST00000497081.2
|
MFAP4
|
microfibrillar-associated protein 4 |
| chr10_-_15413035 | 0.19 |
ENST00000378116.4
ENST00000455654.1 |
FAM171A1
|
family with sequence similarity 171, member A1 |
| chr3_+_183353356 | 0.19 |
ENST00000242810.6
ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24
|
kelch-like family member 24 |
| chr10_+_123970670 | 0.19 |
ENST00000496913.2
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr1_-_161279749 | 0.19 |
ENST00000533357.1
ENST00000360451.6 ENST00000336559.4 |
MPZ
|
myelin protein zero |
| chr5_-_157002775 | 0.19 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr11_+_107461804 | 0.18 |
ENST00000531234.1
|
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr1_+_101185290 | 0.18 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr2_+_232260254 | 0.18 |
ENST00000287590.5
|
B3GNT7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr12_-_49463753 | 0.18 |
ENST00000301068.6
|
RHEBL1
|
Ras homolog enriched in brain like 1 |
| chr19_-_41222775 | 0.18 |
ENST00000324464.3
ENST00000450541.1 ENST00000594720.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr17_-_42908155 | 0.18 |
ENST00000426548.1
ENST00000590758.1 ENST00000591424.1 |
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr11_-_93271058 | 0.17 |
ENST00000527149.1
|
SMCO4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr1_+_145293114 | 0.17 |
ENST00000369338.1
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr5_-_147211226 | 0.17 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr8_+_77593448 | 0.17 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr1_-_1182102 | 0.17 |
ENST00000330388.2
|
FAM132A
|
family with sequence similarity 132, member A |
| chr16_-_3149278 | 0.17 |
ENST00000575108.1
ENST00000576483.1 ENST00000538082.2 ENST00000576985.1 |
ZSCAN10
|
zinc finger and SCAN domain containing 10 |
| chr7_-_100034060 | 0.17 |
ENST00000292330.2
|
PPP1R35
|
protein phosphatase 1, regulatory subunit 35 |
| chr1_+_197871740 | 0.17 |
ENST00000367393.3
|
C1orf53
|
chromosome 1 open reading frame 53 |
| chr22_+_29702572 | 0.17 |
ENST00000407647.2
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr1_+_113217073 | 0.16 |
ENST00000369645.1
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr14_-_105420241 | 0.16 |
ENST00000557457.1
|
AHNAK2
|
AHNAK nucleoprotein 2 |
| chr17_-_4643114 | 0.16 |
ENST00000293778.6
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr3_+_158991025 | 0.16 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr19_+_41725140 | 0.16 |
ENST00000359092.3
|
AXL
|
AXL receptor tyrosine kinase |
| chr22_-_37584321 | 0.16 |
ENST00000397110.2
ENST00000337843.2 |
C1QTNF6
|
C1q and tumor necrosis factor related protein 6 |
| chrX_+_52926322 | 0.16 |
ENST00000430150.2
ENST00000452021.2 ENST00000412319.1 |
FAM156B
|
family with sequence similarity 156, member B |
| chr4_+_183370146 | 0.16 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr3_-_46608010 | 0.16 |
ENST00000395905.3
|
LRRC2
|
leucine rich repeat containing 2 |
| chr9_+_117904097 | 0.16 |
ENST00000374016.1
|
DEC1
|
deleted in esophageal cancer 1 |
| chr1_+_181003067 | 0.16 |
ENST00000434571.2
ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1
|
major histocompatibility complex, class I-related |
| chr10_-_99030395 | 0.16 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr5_-_78281623 | 0.15 |
ENST00000521117.1
|
ARSB
|
arylsulfatase B |
| chr5_-_78281603 | 0.15 |
ENST00000264914.4
|
ARSB
|
arylsulfatase B |
| chr22_-_26879734 | 0.15 |
ENST00000422379.2
ENST00000336873.5 ENST00000398145.2 |
HPS4
|
Hermansky-Pudlak syndrome 4 |
| chr14_+_24590560 | 0.15 |
ENST00000558325.1
|
RP11-468E2.6
|
RP11-468E2.6 |
| chr6_-_28226984 | 0.15 |
ENST00000423974.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr16_-_30798492 | 0.15 |
ENST00000262525.4
|
ZNF629
|
zinc finger protein 629 |
| chr17_+_75369400 | 0.15 |
ENST00000590059.1
|
SEPT9
|
septin 9 |
| chr10_-_61513146 | 0.15 |
ENST00000430431.1
|
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr16_+_11439286 | 0.15 |
ENST00000312499.5
ENST00000576027.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr4_+_129730839 | 0.15 |
ENST00000511647.1
|
PHF17
|
jade family PHD finger 1 |
| chr6_-_16761678 | 0.15 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr2_+_177053307 | 0.15 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr3_+_46616017 | 0.15 |
ENST00000542931.1
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr11_-_59633951 | 0.15 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr14_-_21562648 | 0.14 |
ENST00000555270.1
|
ZNF219
|
zinc finger protein 219 |
| chr14_-_23772032 | 0.14 |
ENST00000452015.4
|
PPP1R3E
|
protein phosphatase 1, regulatory subunit 3E |
| chr17_-_38210644 | 0.14 |
ENST00000394128.2
ENST00000394127.2 ENST00000356271.3 ENST00000535071.2 ENST00000580885.1 ENST00000543759.2 ENST00000537674.2 ENST00000580517.1 ENST00000578161.1 |
MED24
|
mediator complex subunit 24 |
| chr12_+_10460417 | 0.14 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr11_-_86383157 | 0.14 |
ENST00000393324.3
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr16_-_57570450 | 0.14 |
ENST00000258214.2
|
CCDC102A
|
coiled-coil domain containing 102A |
| chr7_-_141401951 | 0.14 |
ENST00000536163.1
|
KIAA1147
|
KIAA1147 |
| chr3_-_98235962 | 0.13 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr16_+_3068393 | 0.13 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr1_+_202431859 | 0.13 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr5_+_125758813 | 0.13 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chrX_-_100307076 | 0.13 |
ENST00000338687.7
ENST00000545398.1 ENST00000372931.5 |
TRMT2B
|
tRNA methyltransferase 2 homolog B (S. cerevisiae) |
| chr4_-_141074123 | 0.13 |
ENST00000502696.1
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr9_+_129097520 | 0.13 |
ENST00000436593.3
|
MVB12B
|
multivesicular body subunit 12B |
| chr6_-_34524093 | 0.13 |
ENST00000544425.1
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr16_-_75285380 | 0.13 |
ENST00000393420.6
ENST00000162330.5 |
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr9_+_129097479 | 0.13 |
ENST00000402437.2
|
MVB12B
|
multivesicular body subunit 12B |
| chr8_+_118532937 | 0.13 |
ENST00000297347.3
|
MED30
|
mediator complex subunit 30 |
| chr14_-_21566731 | 0.13 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr10_-_28571015 | 0.12 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr12_-_58329819 | 0.12 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr17_-_4852332 | 0.12 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr17_+_44370099 | 0.12 |
ENST00000496930.1
|
LRRC37A
|
leucine rich repeat containing 37A |
| chr9_-_117111222 | 0.12 |
ENST00000374079.4
|
AKNA
|
AT-hook transcription factor |
| chr9_+_74764278 | 0.12 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr3_-_50336278 | 0.12 |
ENST00000359051.3
ENST00000417393.1 ENST00000442620.1 ENST00000452674.1 |
HYAL3
NAT6
|
hyaluronoglucosaminidase 3 N-acetyltransferase 6 (GCN5-related) |
| chr11_+_46354455 | 0.12 |
ENST00000343674.6
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr7_-_137686791 | 0.12 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr1_+_165796753 | 0.12 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr22_-_38577782 | 0.12 |
ENST00000430886.1
ENST00000332509.3 ENST00000447598.2 ENST00000435484.1 ENST00000402064.1 ENST00000436218.1 |
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr19_-_53400813 | 0.12 |
ENST00000595635.1
ENST00000594741.1 ENST00000597111.1 ENST00000593618.1 ENST00000597909.1 |
ZNF320
|
zinc finger protein 320 |
| chr15_+_86087267 | 0.12 |
ENST00000558166.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr9_-_117150243 | 0.12 |
ENST00000374088.3
|
AKNA
|
AT-hook transcription factor |
| chr1_+_12538594 | 0.12 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr11_+_123396307 | 0.12 |
ENST00000456860.2
|
GRAMD1B
|
GRAM domain containing 1B |
| chr3_+_46449049 | 0.12 |
ENST00000357392.4
ENST00000400880.3 ENST00000433848.1 |
CCRL2
|
chemokine (C-C motif) receptor-like 2 |
| chr13_-_38172863 | 0.12 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr3_+_135969148 | 0.11 |
ENST00000251654.4
ENST00000490504.1 ENST00000483687.1 ENST00000468777.1 ENST00000462637.1 ENST00000466072.1 ENST00000482086.1 ENST00000471595.1 ENST00000469217.1 ENST00000465423.1 ENST00000478469.1 |
PCCB
|
propionyl CoA carboxylase, beta polypeptide |
| chr11_-_128894053 | 0.11 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr9_+_107526443 | 0.11 |
ENST00000374762.3
|
NIPSNAP3B
|
nipsnap homolog 3B (C. elegans) |
| chr1_-_156542328 | 0.11 |
ENST00000361170.2
|
IQGAP3
|
IQ motif containing GTPase activating protein 3 |
| chr12_-_27167233 | 0.11 |
ENST00000535819.1
ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3
|
transmembrane 7 superfamily member 3 |
| chr2_+_204192942 | 0.11 |
ENST00000295851.5
ENST00000261017.5 |
ABI2
|
abl-interactor 2 |
| chr1_+_24646263 | 0.11 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr17_-_47755338 | 0.11 |
ENST00000508805.1
ENST00000515508.2 ENST00000451526.2 ENST00000507970.1 |
SPOP
|
speckle-type POZ protein |
| chr3_-_10362725 | 0.11 |
ENST00000397109.3
ENST00000428626.1 ENST00000445064.1 ENST00000431352.1 ENST00000397117.1 ENST00000337354.4 ENST00000383801.2 ENST00000432213.1 ENST00000350697.3 |
SEC13
|
SEC13 homolog (S. cerevisiae) |
| chr14_+_60716276 | 0.11 |
ENST00000528241.2
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr5_+_125758865 | 0.11 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr22_-_39268308 | 0.11 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr19_-_46105411 | 0.11 |
ENST00000323040.4
ENST00000544371.1 |
GPR4
OPA3
|
G protein-coupled receptor 4 optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr19_-_14316980 | 0.11 |
ENST00000361434.3
ENST00000340736.6 |
LPHN1
|
latrophilin 1 |
| chr22_+_31608219 | 0.10 |
ENST00000406516.1
ENST00000444929.2 ENST00000331728.4 |
LIMK2
|
LIM domain kinase 2 |
| chr1_+_113217043 | 0.10 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr17_-_27188984 | 0.10 |
ENST00000582320.2
|
MIR144
|
microRNA 451b |
| chr10_+_5090940 | 0.10 |
ENST00000602997.1
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr3_-_196159268 | 0.10 |
ENST00000381887.3
ENST00000535858.1 ENST00000428095.1 ENST00000296328.4 |
UBXN7
|
UBX domain protein 7 |
| chr1_-_221915418 | 0.10 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr22_+_31892373 | 0.10 |
ENST00000443011.1
ENST00000400289.1 ENST00000444859.1 ENST00000400288.2 |
SFI1
|
Sfi1 homolog, spindle assembly associated (yeast) |
| chr16_+_79747740 | 0.10 |
ENST00000561510.1
|
RP11-345M22.1
|
RP11-345M22.1 |
| chr2_-_112614424 | 0.10 |
ENST00000427997.1
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr20_+_30946106 | 0.10 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr22_+_42229100 | 0.10 |
ENST00000361204.4
|
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr17_-_1029052 | 0.10 |
ENST00000574437.1
|
ABR
|
active BCR-related |
| chr2_-_208994548 | 0.10 |
ENST00000282141.3
|
CRYGC
|
crystallin, gamma C |
| chr13_-_81801115 | 0.10 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF652
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.2 | 1.1 | GO:1903284 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.7 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.1 | 0.4 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 1.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 0.2 | GO:0060164 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) regulation of timing of neuron differentiation(GO:0060164) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.1 | 0.5 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.4 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.2 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 | 0.2 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.1 | 0.2 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.3 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.6 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.7 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.3 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.0 | 0.2 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.3 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.5 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.3 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 0.3 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.2 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.3 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.3 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.2 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.4 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.6 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.7 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.4 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.0 | GO:0072716 | response to actinomycin D(GO:0072716) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) proline transmembrane transport(GO:0035524) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 1.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.2 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.2 | 1.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 1.1 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.4 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.8 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.3 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.2 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.3 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 0.2 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.1 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.2 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.4 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.6 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |