Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
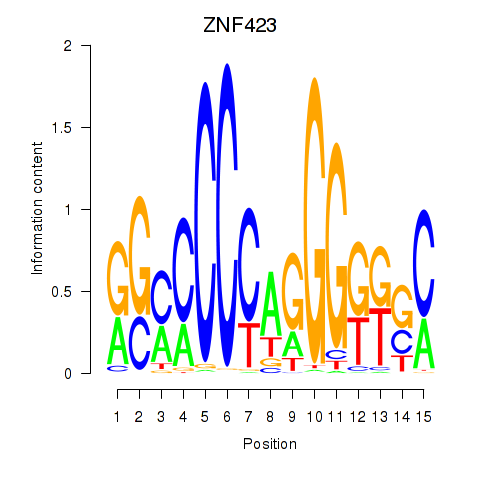
Results for ZNF423
Z-value: 0.45
Transcription factors associated with ZNF423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF423
|
ENSG00000102935.7 | zinc finger protein 423 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF423 | hg19_v2_chr16_-_49891694_49891830 | -0.36 | 4.9e-01 | Click! |
Activity profile of ZNF423 motif
Sorted Z-values of ZNF423 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_33663474 | 0.40 |
ENST00000594414.1
|
SBP1
|
SBP1; Uncharacterized protein |
| chr16_+_27324983 | 0.25 |
ENST00000566117.1
|
IL4R
|
interleukin 4 receptor |
| chr16_-_2162831 | 0.19 |
ENST00000483024.1
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr18_-_60985914 | 0.18 |
ENST00000589955.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr15_-_41522889 | 0.16 |
ENST00000458580.2
ENST00000314992.5 ENST00000558396.1 |
EXD1
|
exonuclease 3'-5' domain containing 1 |
| chr12_+_7342271 | 0.16 |
ENST00000434354.2
ENST00000544456.1 ENST00000545574.1 ENST00000420616.2 |
PEX5
|
peroxisomal biogenesis factor 5 |
| chr5_+_72509751 | 0.15 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr1_-_204121298 | 0.14 |
ENST00000367199.2
|
ETNK2
|
ethanolamine kinase 2 |
| chr19_-_5292781 | 0.14 |
ENST00000586065.1
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr16_+_71392616 | 0.14 |
ENST00000349553.5
ENST00000302628.4 ENST00000562305.1 |
CALB2
|
calbindin 2 |
| chr19_+_14544099 | 0.13 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr7_+_20370746 | 0.13 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr17_-_79212825 | 0.13 |
ENST00000374769.2
|
ENTHD2
|
ENTH domain containing 2 |
| chr12_+_7342441 | 0.12 |
ENST00000412720.2
ENST00000396637.3 |
PEX5
|
peroxisomal biogenesis factor 5 |
| chr5_+_150400124 | 0.12 |
ENST00000388825.4
ENST00000521650.1 ENST00000517973.1 |
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr2_-_206950996 | 0.11 |
ENST00000414320.1
|
INO80D
|
INO80 complex subunit D |
| chr17_+_36584662 | 0.11 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr7_-_27219632 | 0.11 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr1_+_24118452 | 0.11 |
ENST00000421070.1
|
LYPLA2
|
lysophospholipase II |
| chr8_-_80680078 | 0.11 |
ENST00000337919.5
ENST00000354724.3 |
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr16_+_27325202 | 0.10 |
ENST00000395762.2
ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R
|
interleukin 4 receptor |
| chr15_-_75660919 | 0.10 |
ENST00000569482.1
ENST00000565683.1 ENST00000561615.1 ENST00000563622.1 ENST00000568374.1 ENST00000566256.1 ENST00000267978.5 |
MAN2C1
|
mannosidase, alpha, class 2C, member 1 |
| chr1_+_1222489 | 0.10 |
ENST00000379099.3
|
SCNN1D
|
sodium channel, non-voltage-gated 1, delta subunit |
| chr12_+_52643077 | 0.10 |
ENST00000553310.2
ENST00000544024.1 |
KRT86
|
keratin 86 |
| chr5_+_141303461 | 0.10 |
ENST00000508751.1
|
KIAA0141
|
KIAA0141 |
| chr19_-_56110859 | 0.10 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr5_+_154181816 | 0.09 |
ENST00000518677.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr9_-_35619539 | 0.09 |
ENST00000396757.1
|
CD72
|
CD72 molecule |
| chr8_+_22422749 | 0.09 |
ENST00000523900.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr1_-_155177677 | 0.08 |
ENST00000368378.3
ENST00000541990.1 ENST00000457183.2 |
THBS3
|
thrombospondin 3 |
| chr8_+_22022223 | 0.08 |
ENST00000306385.5
|
BMP1
|
bone morphogenetic protein 1 |
| chr19_+_41770349 | 0.08 |
ENST00000602130.1
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr1_-_204121102 | 0.08 |
ENST00000367202.4
|
ETNK2
|
ethanolamine kinase 2 |
| chr20_+_44462481 | 0.08 |
ENST00000491381.1
ENST00000342644.5 ENST00000372542.1 |
SNX21
|
sorting nexin family member 21 |
| chr8_+_22423219 | 0.08 |
ENST00000523965.1
ENST00000521554.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr17_-_27230035 | 0.08 |
ENST00000378895.4
ENST00000394901.3 |
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr14_+_100204027 | 0.07 |
ENST00000554479.1
ENST00000555145.1 |
EML1
|
echinoderm microtubule associated protein like 1 |
| chr11_-_61659006 | 0.07 |
ENST00000278829.2
|
FADS3
|
fatty acid desaturase 3 |
| chr2_-_120980939 | 0.07 |
ENST00000426077.2
|
TMEM185B
|
transmembrane protein 185B |
| chr11_-_47207950 | 0.07 |
ENST00000298838.6
ENST00000531226.1 ENST00000524509.1 ENST00000528201.1 ENST00000530513.1 |
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr19_-_36391434 | 0.07 |
ENST00000396901.1
ENST00000585925.1 |
NFKBID
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta |
| chr12_-_50101165 | 0.07 |
ENST00000352151.5
ENST00000335154.5 ENST00000293590.5 |
FMNL3
|
formin-like 3 |
| chr11_+_60691924 | 0.07 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr9_-_131486367 | 0.07 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr11_-_504489 | 0.07 |
ENST00000529368.1
|
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr19_+_10531150 | 0.07 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr14_-_93582148 | 0.07 |
ENST00000267615.6
ENST00000553452.1 |
ITPK1
|
inositol-tetrakisphosphate 1-kinase |
| chr1_-_33366931 | 0.07 |
ENST00000373463.3
ENST00000329151.5 |
TMEM54
|
transmembrane protein 54 |
| chr2_+_135011731 | 0.07 |
ENST00000281923.2
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr17_-_1029052 | 0.06 |
ENST00000574437.1
|
ABR
|
active BCR-related |
| chr1_-_35658736 | 0.06 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr8_+_22423168 | 0.06 |
ENST00000518912.1
ENST00000428103.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr19_+_41770269 | 0.06 |
ENST00000378215.4
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr17_-_48227877 | 0.06 |
ENST00000316878.6
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B |
| chr17_+_6347761 | 0.06 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr5_+_141303373 | 0.06 |
ENST00000432126.2
ENST00000194118.4 |
KIAA0141
|
KIAA0141 |
| chr17_+_6347729 | 0.06 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr22_-_37823468 | 0.06 |
ENST00000402918.2
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| chr20_+_43029911 | 0.06 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr12_+_54410664 | 0.06 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr19_-_23941680 | 0.06 |
ENST00000402377.3
|
ZNF681
|
zinc finger protein 681 |
| chr19_-_4066890 | 0.06 |
ENST00000322357.4
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr14_-_93582214 | 0.06 |
ENST00000556603.2
ENST00000354313.3 |
ITPK1
|
inositol-tetrakisphosphate 1-kinase |
| chr9_+_116638562 | 0.05 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr17_+_77020224 | 0.05 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr3_+_9958758 | 0.05 |
ENST00000383812.4
ENST00000438091.1 ENST00000295981.3 ENST00000436503.1 ENST00000403601.3 ENST00000416074.2 ENST00000455057.1 |
IL17RC
|
interleukin 17 receptor C |
| chr6_+_42896865 | 0.05 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr6_+_33589161 | 0.05 |
ENST00000605930.1
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr11_-_6495101 | 0.05 |
ENST00000528227.1
ENST00000359518.3 ENST00000345851.3 ENST00000537602.1 |
TRIM3
|
tripartite motif containing 3 |
| chr22_-_38484922 | 0.05 |
ENST00000428572.1
|
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr17_+_77020146 | 0.05 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr7_+_101460882 | 0.05 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr17_+_77020325 | 0.05 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr8_+_22022653 | 0.05 |
ENST00000354870.5
ENST00000397816.3 ENST00000306349.8 |
BMP1
|
bone morphogenetic protein 1 |
| chr10_-_118032979 | 0.05 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr7_-_30029328 | 0.05 |
ENST00000425819.2
ENST00000409570.1 |
SCRN1
|
secernin 1 |
| chr12_-_125348448 | 0.04 |
ENST00000339570.5
|
SCARB1
|
scavenger receptor class B, member 1 |
| chr20_-_36794938 | 0.04 |
ENST00000453095.1
|
TGM2
|
transglutaminase 2 |
| chr8_+_145065521 | 0.04 |
ENST00000534791.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr12_-_125348329 | 0.04 |
ENST00000546215.1
ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1
|
scavenger receptor class B, member 1 |
| chr20_+_44462430 | 0.04 |
ENST00000462307.1
|
SNX21
|
sorting nexin family member 21 |
| chr9_-_130659569 | 0.04 |
ENST00000542456.1
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr6_+_151187074 | 0.04 |
ENST00000367308.4
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr7_-_5570229 | 0.04 |
ENST00000331789.5
|
ACTB
|
actin, beta |
| chr1_-_204121013 | 0.04 |
ENST00000367201.3
|
ETNK2
|
ethanolamine kinase 2 |
| chr11_+_73019282 | 0.04 |
ENST00000263674.3
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr20_-_36794902 | 0.04 |
ENST00000373403.3
|
TGM2
|
transglutaminase 2 |
| chr3_+_196439275 | 0.04 |
ENST00000296333.5
|
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr19_-_4043154 | 0.04 |
ENST00000535853.1
|
AC016586.1
|
AC016586.1 |
| chr12_-_50101003 | 0.04 |
ENST00000550488.1
|
FMNL3
|
formin-like 3 |
| chr19_-_10450287 | 0.04 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr19_-_10679644 | 0.04 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr3_-_196439065 | 0.04 |
ENST00000399942.4
ENST00000409690.3 |
CEP19
|
centrosomal protein 19kDa |
| chr9_-_130742792 | 0.04 |
ENST00000373095.1
|
FAM102A
|
family with sequence similarity 102, member A |
| chr9_+_131549610 | 0.04 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr8_+_22022800 | 0.04 |
ENST00000397814.3
|
BMP1
|
bone morphogenetic protein 1 |
| chr8_+_30241934 | 0.04 |
ENST00000538486.1
|
RBPMS
|
RNA binding protein with multiple splicing |
| chr3_+_9958870 | 0.04 |
ENST00000413608.1
|
IL17RC
|
interleukin 17 receptor C |
| chr3_-_52931557 | 0.04 |
ENST00000504329.1
ENST00000355083.5 |
TMEM110-MUSTN1
TMEM110
|
TMEM110-MUSTN1 readthrough transmembrane protein 110 |
| chr11_-_44972418 | 0.03 |
ENST00000525680.1
ENST00000528290.1 ENST00000530035.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr9_-_130659679 | 0.03 |
ENST00000373141.1
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr8_+_30241995 | 0.03 |
ENST00000397323.4
ENST00000339877.4 ENST00000320203.4 ENST00000287771.5 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr11_-_44971702 | 0.03 |
ENST00000533940.1
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr6_-_143266297 | 0.03 |
ENST00000367603.2
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr17_+_7341586 | 0.03 |
ENST00000575235.1
|
FGF11
|
fibroblast growth factor 11 |
| chr1_+_17914907 | 0.03 |
ENST00000375420.3
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr19_-_38714847 | 0.03 |
ENST00000420980.2
ENST00000355526.4 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr12_+_7037461 | 0.03 |
ENST00000396684.2
|
ATN1
|
atrophin 1 |
| chr20_+_44462749 | 0.03 |
ENST00000372541.1
|
SNX21
|
sorting nexin family member 21 |
| chr2_-_46769694 | 0.03 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr19_-_10450328 | 0.03 |
ENST00000160262.5
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr16_-_11681023 | 0.03 |
ENST00000570904.1
ENST00000574701.1 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr19_-_11689803 | 0.03 |
ENST00000591319.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr11_-_6495082 | 0.03 |
ENST00000536344.1
|
TRIM3
|
tripartite motif containing 3 |
| chr19_-_15311713 | 0.03 |
ENST00000601011.1
ENST00000263388.2 |
NOTCH3
|
notch 3 |
| chr19_+_41770236 | 0.03 |
ENST00000392006.3
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr7_-_27219849 | 0.02 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr22_-_43045574 | 0.02 |
ENST00000352397.5
|
CYB5R3
|
cytochrome b5 reductase 3 |
| chr12_-_57522813 | 0.02 |
ENST00000556155.1
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr21_+_45432174 | 0.02 |
ENST00000380221.3
ENST00000291574.4 |
TRAPPC10
|
trafficking protein particle complex 10 |
| chr7_-_100491854 | 0.02 |
ENST00000426415.1
ENST00000430554.1 ENST00000412389.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr11_-_67120974 | 0.02 |
ENST00000539074.1
ENST00000312419.3 |
POLD4
|
polymerase (DNA-directed), delta 4, accessory subunit |
| chr10_-_100027943 | 0.02 |
ENST00000260702.3
|
LOXL4
|
lysyl oxidase-like 4 |
| chr11_-_44972299 | 0.02 |
ENST00000528473.1
|
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr13_-_46626847 | 0.02 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr22_-_31742218 | 0.02 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr9_+_130160440 | 0.02 |
ENST00000439597.1
ENST00000423934.1 |
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr10_+_88428370 | 0.02 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr19_+_55795493 | 0.02 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr16_+_70148230 | 0.02 |
ENST00000398122.3
ENST00000568530.1 |
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit |
| chr9_-_16870704 | 0.02 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chr6_-_35480640 | 0.02 |
ENST00000428978.1
ENST00000322263.4 |
TULP1
|
tubby like protein 1 |
| chr4_-_120548779 | 0.02 |
ENST00000264805.5
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr15_+_41523417 | 0.02 |
ENST00000560397.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr9_+_112542591 | 0.02 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr8_+_145065705 | 0.02 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr5_-_176057365 | 0.02 |
ENST00000310112.3
|
SNCB
|
synuclein, beta |
| chr9_-_39239171 | 0.01 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr12_-_54779511 | 0.01 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr19_-_57183114 | 0.01 |
ENST00000537055.2
ENST00000601659.1 |
ZNF835
|
zinc finger protein 835 |
| chr5_-_172756506 | 0.01 |
ENST00000265087.4
|
STC2
|
stanniocalcin 2 |
| chr11_-_61658853 | 0.01 |
ENST00000525588.1
ENST00000540820.1 |
FADS3
|
fatty acid desaturase 3 |
| chr12_+_7342178 | 0.01 |
ENST00000266563.5
ENST00000543974.1 |
PEX5
|
peroxisomal biogenesis factor 5 |
| chr9_+_139249228 | 0.01 |
ENST00000392944.1
|
GPSM1
|
G-protein signaling modulator 1 |
| chr7_+_150748288 | 0.01 |
ENST00000490540.1
|
ASIC3
|
acid-sensing (proton-gated) ion channel 3 |
| chr1_-_203055129 | 0.01 |
ENST00000241651.4
|
MYOG
|
myogenin (myogenic factor 4) |
| chr10_-_121296045 | 0.01 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr6_+_30130969 | 0.01 |
ENST00000376694.4
|
TRIM15
|
tripartite motif containing 15 |
| chr19_+_45582453 | 0.01 |
ENST00000591607.1
ENST00000591747.1 ENST00000270257.4 ENST00000391951.2 ENST00000587566.1 |
GEMIN7
MARK4
|
gem (nuclear organelle) associated protein 7 MAP/microtubule affinity-regulating kinase 4 |
| chr21_-_18985230 | 0.01 |
ENST00000457956.1
ENST00000348354.6 |
BTG3
|
BTG family, member 3 |
| chr6_+_151186554 | 0.01 |
ENST00000367321.3
ENST00000367307.4 |
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr5_+_135385202 | 0.01 |
ENST00000514554.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr10_+_17271266 | 0.01 |
ENST00000224237.5
|
VIM
|
vimentin |
| chr16_+_30675654 | 0.00 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr22_+_35653445 | 0.00 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr3_+_196439170 | 0.00 |
ENST00000392391.3
ENST00000314118.4 |
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr4_+_74735102 | 0.00 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr12_-_118810688 | 0.00 |
ENST00000542532.1
ENST00000392533.3 |
TAOK3
|
TAO kinase 3 |
| chrX_-_1572629 | 0.00 |
ENST00000534940.1
|
ASMTL
|
acetylserotonin O-methyltransferase-like |
| chr15_-_59225844 | 0.00 |
ENST00000380516.2
|
SLTM
|
SAFB-like, transcription modulator |
| chr16_+_3014269 | 0.00 |
ENST00000575885.1
ENST00000571007.1 ENST00000319500.6 |
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr1_+_155278625 | 0.00 |
ENST00000368356.4
ENST00000356657.6 |
FDPS
|
farnesyl diphosphate synthase |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF423
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.1 | 0.3 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.2 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.2 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.2 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:0052825 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |