Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
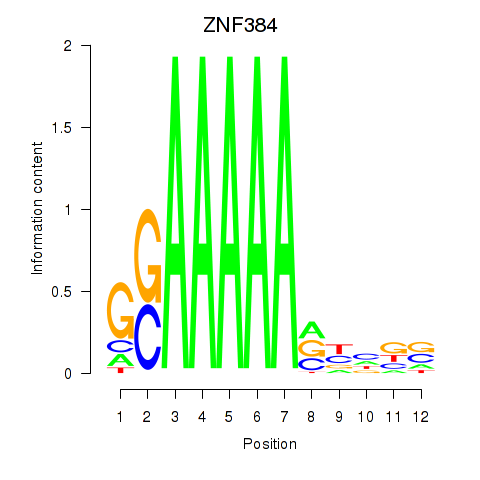
Results for ZNF384
Z-value: 1.68
Transcription factors associated with ZNF384
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF384
|
ENSG00000126746.13 | zinc finger protein 384 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF384 | hg19_v2_chr12_-_6798616_6798684 | -0.98 | 4.3e-04 | Click! |
Activity profile of ZNF384 motif
Sorted Z-values of ZNF384 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_91152303 | 3.52 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr10_+_91087651 | 1.13 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr1_-_63988846 | 1.11 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr12_+_113344755 | 1.07 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr12_+_113344582 | 0.98 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chrX_+_122993544 | 0.96 |
ENST00000422098.1
|
XIAP
|
X-linked inhibitor of apoptosis |
| chr12_+_100594557 | 0.94 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr2_+_187371440 | 0.90 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr12_-_76742183 | 0.87 |
ENST00000393262.3
|
BBS10
|
Bardet-Biedl syndrome 10 |
| chr4_+_78804393 | 0.83 |
ENST00000502384.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr3_-_185641681 | 0.83 |
ENST00000259043.7
|
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr14_+_94577074 | 0.80 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr3_+_172034218 | 0.79 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr2_+_38177575 | 0.78 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr8_-_90993869 | 0.76 |
ENST00000517772.1
|
NBN
|
nibrin |
| chr5_+_118691008 | 0.74 |
ENST00000504642.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr14_-_51027838 | 0.74 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr7_-_54826869 | 0.74 |
ENST00000450622.1
|
SEC61G
|
Sec61 gamma subunit |
| chr2_-_170430277 | 0.73 |
ENST00000438035.1
ENST00000453929.2 |
FASTKD1
|
FAST kinase domains 1 |
| chr1_+_97187318 | 0.70 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr1_-_193075180 | 0.70 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr4_+_89299994 | 0.68 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr9_-_127952187 | 0.67 |
ENST00000451402.1
ENST00000415905.1 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr1_+_65886326 | 0.67 |
ENST00000371059.3
ENST00000371060.3 ENST00000349533.6 ENST00000406510.3 |
LEPR
|
leptin receptor |
| chr12_-_42631529 | 0.65 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr20_+_52824367 | 0.65 |
ENST00000371419.2
|
PFDN4
|
prefoldin subunit 4 |
| chr1_+_156308403 | 0.65 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chrX_-_108976521 | 0.65 |
ENST00000469796.2
ENST00000502391.1 ENST00000508092.1 ENST00000340800.2 ENST00000348502.6 |
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr3_-_112738490 | 0.65 |
ENST00000393857.2
|
C3orf17
|
chromosome 3 open reading frame 17 |
| chr12_+_28299014 | 0.64 |
ENST00000538586.1
ENST00000536154.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr6_+_46620676 | 0.64 |
ENST00000371347.5
ENST00000411689.2 |
SLC25A27
|
solute carrier family 25, member 27 |
| chr12_-_75905374 | 0.64 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr6_-_110012380 | 0.64 |
ENST00000424296.2
ENST00000341338.6 ENST00000368948.2 ENST00000285397.5 |
AK9
|
adenylate kinase 9 |
| chr2_+_67624430 | 0.63 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr11_-_88070896 | 0.63 |
ENST00000529974.1
ENST00000527018.1 |
CTSC
|
cathepsin C |
| chr4_+_89299885 | 0.62 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr3_+_45730829 | 0.62 |
ENST00000438671.1
|
SACM1L
|
SAC1 suppressor of actin mutations 1-like (yeast) |
| chr14_+_24630465 | 0.61 |
ENST00000557894.1
ENST00000559284.1 ENST00000560275.1 |
IRF9
|
interferon regulatory factor 9 |
| chr2_+_114163945 | 0.60 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr10_+_126490354 | 0.60 |
ENST00000298492.5
|
FAM175B
|
family with sequence similarity 175, member B |
| chr4_+_103790462 | 0.60 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr6_-_100016678 | 0.59 |
ENST00000523799.1
ENST00000520429.1 |
CCNC
|
cyclin C |
| chr14_+_101361107 | 0.59 |
ENST00000553584.1
ENST00000554852.1 |
MEG8
|
maternally expressed 8 (non-protein coding) |
| chr11_-_615942 | 0.58 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr6_-_137540477 | 0.58 |
ENST00000367735.2
ENST00000367739.4 ENST00000458076.1 ENST00000414770.1 |
IFNGR1
|
interferon gamma receptor 1 |
| chr12_-_69080590 | 0.57 |
ENST00000433116.2
ENST00000500695.2 |
RP11-637A17.2
|
RP11-637A17.2 |
| chr12_-_7245018 | 0.56 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr12_-_7245080 | 0.56 |
ENST00000541042.1
ENST00000540242.1 |
C1R
|
complement component 1, r subcomponent |
| chr3_-_108308241 | 0.56 |
ENST00000295746.8
|
KIAA1524
|
KIAA1524 |
| chr8_+_142264664 | 0.55 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chr4_-_492891 | 0.55 |
ENST00000338977.5
ENST00000511833.2 |
ZNF721
|
zinc finger protein 721 |
| chr12_-_7245152 | 0.55 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr6_-_18249971 | 0.54 |
ENST00000507591.1
|
DEK
|
DEK oncogene |
| chr1_+_150039877 | 0.54 |
ENST00000419023.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr12_+_69753448 | 0.54 |
ENST00000247843.2
ENST00000548020.1 ENST00000549685.1 ENST00000552955.1 |
YEATS4
|
YEATS domain containing 4 |
| chr1_-_193074504 | 0.54 |
ENST00000367439.3
|
GLRX2
|
glutaredoxin 2 |
| chr15_-_35280426 | 0.53 |
ENST00000559564.1
ENST00000356321.4 |
ZNF770
|
zinc finger protein 770 |
| chr14_-_64108125 | 0.53 |
ENST00000267522.3
|
WDR89
|
WD repeat domain 89 |
| chr16_-_90096309 | 0.53 |
ENST00000408886.2
|
C16orf3
|
chromosome 16 open reading frame 3 |
| chr20_-_13765526 | 0.53 |
ENST00000202816.1
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr6_+_46620705 | 0.53 |
ENST00000452689.2
|
SLC25A27
|
solute carrier family 25, member 27 |
| chr5_+_118690466 | 0.53 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr18_-_53303123 | 0.52 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr2_+_109403193 | 0.52 |
ENST00000412964.2
ENST00000295124.4 |
CCDC138
|
coiled-coil domain containing 138 |
| chr5_-_145214893 | 0.52 |
ENST00000394450.2
|
PRELID2
|
PRELI domain containing 2 |
| chr4_+_164265035 | 0.52 |
ENST00000338566.3
|
NPY5R
|
neuropeptide Y receptor Y5 |
| chrX_-_135056106 | 0.52 |
ENST00000433339.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr14_+_45553296 | 0.51 |
ENST00000355765.6
ENST00000553605.1 |
PRPF39
|
pre-mRNA processing factor 39 |
| chr10_-_126849626 | 0.51 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr4_+_26323764 | 0.51 |
ENST00000514730.1
ENST00000507574.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr6_-_46620522 | 0.51 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr12_+_28410128 | 0.50 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr4_-_15661474 | 0.50 |
ENST00000509314.1
ENST00000503196.1 |
FBXL5
|
F-box and leucine-rich repeat protein 5 |
| chr15_+_49913201 | 0.50 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr11_+_102217936 | 0.50 |
ENST00000532832.1
ENST00000530675.1 ENST00000533742.1 ENST00000227758.2 ENST00000532672.1 ENST00000531259.1 ENST00000527465.1 |
BIRC2
|
baculoviral IAP repeat containing 2 |
| chr8_-_40200903 | 0.50 |
ENST00000522486.1
|
CTA-392C11.1
|
CTA-392C11.1 |
| chr4_-_153700864 | 0.49 |
ENST00000304337.2
|
TIGD4
|
tigger transposable element derived 4 |
| chr10_+_25305524 | 0.49 |
ENST00000524413.1
ENST00000376356.4 |
THNSL1
|
threonine synthase-like 1 (S. cerevisiae) |
| chr12_-_7245125 | 0.49 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr1_-_235098935 | 0.48 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr19_-_17571722 | 0.48 |
ENST00000301944.2
|
NXNL1
|
nucleoredoxin-like 1 |
| chr3_-_122283079 | 0.48 |
ENST00000471785.1
ENST00000466126.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr15_+_48413169 | 0.48 |
ENST00000341459.3
ENST00000482911.2 |
SLC24A5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chrX_-_55187588 | 0.48 |
ENST00000472571.2
ENST00000332132.4 ENST00000425133.2 ENST00000358460.4 |
FAM104B
|
family with sequence similarity 104, member B |
| chrX_-_135056216 | 0.48 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr1_+_10057274 | 0.47 |
ENST00000294435.7
|
RBP7
|
retinol binding protein 7, cellular |
| chr3_-_58523010 | 0.47 |
ENST00000459701.2
ENST00000302819.5 |
ACOX2
|
acyl-CoA oxidase 2, branched chain |
| chr15_-_35261996 | 0.47 |
ENST00000156471.5
|
AQR
|
aquarius intron-binding spliceosomal factor |
| chr2_-_188419078 | 0.47 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr2_-_44588624 | 0.47 |
ENST00000438314.1
ENST00000409936.1 |
PREPL
|
prolyl endopeptidase-like |
| chr3_-_122283100 | 0.47 |
ENST00000492382.1
ENST00000462315.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr7_+_45927956 | 0.47 |
ENST00000275525.3
ENST00000457280.1 |
IGFBP1
|
insulin-like growth factor binding protein 1 |
| chr15_+_48413211 | 0.46 |
ENST00000449382.2
|
SLC24A5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chr21_-_18985230 | 0.46 |
ENST00000457956.1
ENST00000348354.6 |
BTG3
|
BTG family, member 3 |
| chr7_-_103848405 | 0.46 |
ENST00000447452.2
ENST00000545943.1 ENST00000297431.4 |
ORC5
|
origin recognition complex, subunit 5 |
| chr8_+_30013875 | 0.46 |
ENST00000520829.1
|
DCTN6
|
dynactin 6 |
| chr2_+_190722119 | 0.46 |
ENST00000452382.1
|
PMS1
|
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| chr12_-_100486668 | 0.46 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr1_-_85156417 | 0.45 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr13_-_20767037 | 0.45 |
ENST00000382848.4
|
GJB2
|
gap junction protein, beta 2, 26kDa |
| chr2_-_99797390 | 0.45 |
ENST00000422537.2
|
MITD1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr4_-_39034542 | 0.45 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr18_-_64271316 | 0.45 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chrX_-_108976410 | 0.45 |
ENST00000504980.1
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr7_-_35013217 | 0.45 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr11_-_88070920 | 0.45 |
ENST00000524463.1
ENST00000227266.5 |
CTSC
|
cathepsin C |
| chr2_+_172778952 | 0.45 |
ENST00000392584.1
ENST00000264108.4 |
HAT1
|
histone acetyltransferase 1 |
| chr10_-_96122682 | 0.45 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr2_-_170430366 | 0.45 |
ENST00000453153.2
ENST00000445210.1 |
FASTKD1
|
FAST kinase domains 1 |
| chr7_-_130080681 | 0.44 |
ENST00000469826.1
|
CEP41
|
centrosomal protein 41kDa |
| chr6_+_64346386 | 0.44 |
ENST00000509330.1
|
PHF3
|
PHD finger protein 3 |
| chrX_+_40440146 | 0.44 |
ENST00000535539.1
ENST00000378438.4 ENST00000436783.1 ENST00000544975.1 ENST00000535777.1 ENST00000447485.1 ENST00000423649.1 |
ATP6AP2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr8_+_48920960 | 0.43 |
ENST00000523111.2
ENST00000523432.1 ENST00000521346.1 ENST00000517630.1 |
UBE2V2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr2_+_64068844 | 0.43 |
ENST00000337130.5
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr6_-_116989916 | 0.43 |
ENST00000368576.3
ENST00000368573.1 |
ZUFSP
|
zinc finger with UFM1-specific peptidase domain |
| chr12_+_113354341 | 0.43 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr17_-_61045902 | 0.43 |
ENST00000581596.1
|
RP11-180P8.3
|
RP11-180P8.3 |
| chr17_-_38574169 | 0.43 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr4_-_155533787 | 0.43 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr1_-_94344754 | 0.43 |
ENST00000436063.2
|
DNTTIP2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr6_-_42185583 | 0.43 |
ENST00000053468.3
|
MRPS10
|
mitochondrial ribosomal protein S10 |
| chr19_-_12551849 | 0.43 |
ENST00000595562.1
ENST00000301547.5 |
CTD-3105H18.16
ZNF443
|
Uncharacterized protein zinc finger protein 443 |
| chr11_-_82681626 | 0.43 |
ENST00000534396.1
|
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr1_+_79115503 | 0.43 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr19_+_21203426 | 0.42 |
ENST00000261560.5
ENST00000599548.1 ENST00000594110.1 |
ZNF430
|
zinc finger protein 430 |
| chr8_-_27695552 | 0.42 |
ENST00000522944.1
ENST00000301905.4 |
PBK
|
PDZ binding kinase |
| chr1_+_84944926 | 0.42 |
ENST00000370656.1
ENST00000370654.5 |
RPF1
|
ribosome production factor 1 homolog (S. cerevisiae) |
| chr1_+_150254936 | 0.42 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr1_+_225140372 | 0.42 |
ENST00000366848.1
ENST00000439375.2 |
DNAH14
|
dynein, axonemal, heavy chain 14 |
| chr5_-_38845812 | 0.41 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr7_+_90032667 | 0.41 |
ENST00000496677.1
ENST00000287916.4 ENST00000535571.1 ENST00000394604.1 ENST00000394605.2 |
CLDN12
|
claudin 12 |
| chr15_+_82555169 | 0.41 |
ENST00000565432.1
ENST00000427381.2 |
FAM154B
|
family with sequence similarity 154, member B |
| chr5_+_64920826 | 0.41 |
ENST00000438419.2
ENST00000231526.4 ENST00000505553.1 ENST00000545191.1 |
TRAPPC13
|
trafficking protein particle complex 13 |
| chr19_-_46627914 | 0.41 |
ENST00000341415.2
|
IGFL3
|
IGF-like family member 3 |
| chr7_-_108097144 | 0.41 |
ENST00000418239.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr7_+_106415457 | 0.41 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr21_-_35773370 | 0.41 |
ENST00000410005.1
|
AP000322.54
|
chromosome 21 open reading frame 140 |
| chr2_+_29353520 | 0.40 |
ENST00000438819.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr1_-_77685084 | 0.40 |
ENST00000370812.3
ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr12_+_86268065 | 0.40 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chr7_+_87505544 | 0.40 |
ENST00000265728.1
|
DBF4
|
DBF4 homolog (S. cerevisiae) |
| chr4_+_9446156 | 0.40 |
ENST00000334879.1
|
DEFB131
|
defensin, beta 131 |
| chr3_-_148939598 | 0.40 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr16_+_48657361 | 0.39 |
ENST00000565072.1
|
RP11-42I10.1
|
RP11-42I10.1 |
| chr17_-_64216748 | 0.39 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr2_+_214149113 | 0.39 |
ENST00000331683.5
ENST00000432529.2 ENST00000413312.1 ENST00000272898.7 ENST00000447990.1 |
SPAG16
|
sperm associated antigen 16 |
| chr4_+_178230985 | 0.39 |
ENST00000264596.3
|
NEIL3
|
nei endonuclease VIII-like 3 (E. coli) |
| chr1_-_150693318 | 0.39 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr2_+_183989157 | 0.39 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr8_+_29952914 | 0.39 |
ENST00000321250.8
ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr15_+_47631257 | 0.39 |
ENST00000560636.1
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr15_+_83776324 | 0.39 |
ENST00000379390.6
ENST00000379386.4 ENST00000565774.1 ENST00000565982.1 |
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr2_+_169659121 | 0.39 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr1_+_215740709 | 0.39 |
ENST00000259154.4
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr5_+_112227311 | 0.39 |
ENST00000391338.1
|
ZRSR1
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr16_+_75182376 | 0.39 |
ENST00000570010.1
ENST00000568079.1 ENST00000464850.1 ENST00000332307.4 ENST00000393430.2 |
ZFP1
|
ZFP1 zinc finger protein |
| chr6_+_151773583 | 0.39 |
ENST00000545879.1
|
C6orf211
|
chromosome 6 open reading frame 211 |
| chr12_-_48099754 | 0.39 |
ENST00000380650.4
|
RPAP3
|
RNA polymerase II associated protein 3 |
| chr3_+_101292939 | 0.39 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr19_+_10197463 | 0.38 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr1_+_186798073 | 0.38 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr14_-_50506589 | 0.38 |
ENST00000553914.2
|
RP11-58E21.3
|
RP11-58E21.3 |
| chr7_-_87505658 | 0.38 |
ENST00000341119.5
|
SLC25A40
|
solute carrier family 25, member 40 |
| chr14_-_23292596 | 0.38 |
ENST00000554741.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr13_+_38923959 | 0.38 |
ENST00000379649.1
ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1
|
ubiquitin-fold modifier 1 |
| chr4_-_164253738 | 0.38 |
ENST00000509586.1
ENST00000504391.1 ENST00000512819.1 |
NPY1R
|
neuropeptide Y receptor Y1 |
| chr12_+_100660909 | 0.38 |
ENST00000549687.1
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr2_-_188419200 | 0.37 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr6_+_24775153 | 0.37 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr6_+_12008986 | 0.37 |
ENST00000491710.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr13_+_32889605 | 0.37 |
ENST00000380152.3
ENST00000544455.1 ENST00000530893.2 |
BRCA2
|
breast cancer 2, early onset |
| chr17_-_16557128 | 0.37 |
ENST00000423860.2
ENST00000311331.7 ENST00000583766.1 |
ZNF624
|
zinc finger protein 624 |
| chr15_-_55489097 | 0.37 |
ENST00000260443.4
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr3_+_100211412 | 0.37 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr7_-_81635106 | 0.37 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr4_+_14113592 | 0.37 |
ENST00000502759.1
ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085
|
long intergenic non-protein coding RNA 1085 |
| chr8_-_90996459 | 0.37 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr14_-_64108537 | 0.37 |
ENST00000554717.1
ENST00000394942.2 |
WDR89
|
WD repeat domain 89 |
| chr1_-_101360331 | 0.37 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr5_+_68530668 | 0.37 |
ENST00000506563.1
|
CDK7
|
cyclin-dependent kinase 7 |
| chr4_-_156297949 | 0.37 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr12_-_110939870 | 0.37 |
ENST00000447578.2
ENST00000546588.1 ENST00000360579.7 ENST00000549970.1 ENST00000549578.1 |
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr19_+_4769117 | 0.37 |
ENST00000540211.1
ENST00000317292.3 ENST00000586721.1 ENST00000592709.1 ENST00000588711.1 ENST00000589639.1 ENST00000591008.1 ENST00000592663.1 ENST00000588758.1 |
MIR7-3HG
|
MIR7-3 host gene (non-protein coding) |
| chr5_+_134074231 | 0.37 |
ENST00000514518.1
|
CAMLG
|
calcium modulating ligand |
| chrX_+_23682379 | 0.36 |
ENST00000379349.1
|
PRDX4
|
peroxiredoxin 4 |
| chr11_+_57365150 | 0.36 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr3_+_136537911 | 0.36 |
ENST00000393079.3
|
SLC35G2
|
solute carrier family 35, member G2 |
| chr19_+_57050317 | 0.36 |
ENST00000301318.3
ENST00000591844.1 |
ZFP28
|
ZFP28 zinc finger protein |
| chr2_-_228244013 | 0.36 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr8_-_95487331 | 0.36 |
ENST00000336148.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr5_+_78908388 | 0.36 |
ENST00000296783.3
|
PAPD4
|
PAP associated domain containing 4 |
| chr15_+_69373184 | 0.36 |
ENST00000558147.1
ENST00000440444.1 |
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr9_+_79792269 | 0.36 |
ENST00000376634.4
ENST00000376636.3 ENST00000360280.3 |
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr8_-_95908902 | 0.36 |
ENST00000520509.1
|
CCNE2
|
cyclin E2 |
| chr4_+_109571740 | 0.36 |
ENST00000361564.4
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr7_+_112120908 | 0.36 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr12_+_69202975 | 0.36 |
ENST00000544561.1
ENST00000393410.1 ENST00000299252.4 ENST00000360430.2 ENST00000517852.1 ENST00000545204.1 ENST00000393413.3 ENST00000350057.5 ENST00000348801.2 ENST00000478070.1 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr13_+_53030107 | 0.36 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr8_+_125860939 | 0.35 |
ENST00000525292.1
ENST00000528090.1 |
LINC00964
|
long intergenic non-protein coding RNA 964 |
| chr2_-_122494487 | 0.35 |
ENST00000451734.1
ENST00000285814.4 |
MKI67IP
|
nucleolar protein interacting with the FHA domain of MKI67 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF384
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.4 | 1.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.3 | 1.0 | GO:0061566 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.3 | 1.2 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.2 | 0.6 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.2 | 0.6 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.2 | 1.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.2 | 0.7 | GO:0001080 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.2 | 0.6 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.2 | 0.5 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.2 | 0.6 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 0.6 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.1 | 1.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.6 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 0.4 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.5 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.4 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.1 | 0.4 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.1 | 0.4 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.5 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.3 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.1 | 1.0 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.2 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.1 | 1.2 | GO:0042262 | DNA protection(GO:0042262) |
| 0.1 | 0.9 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 1.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.3 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.8 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.4 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 0.3 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.1 | 0.3 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.5 | GO:1904884 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.4 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.5 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.1 | 0.4 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.1 | 0.8 | GO:1905068 | arterial endothelial cell fate commitment(GO:0060844) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.3 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.1 | GO:1904429 | regulation of t-circle formation(GO:1904429) |
| 0.1 | 0.1 | GO:1901741 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) positive regulation of syncytium formation by plasma membrane fusion(GO:0060143) regulation of myoblast fusion(GO:1901739) positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.8 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.7 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.5 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.5 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.1 | 0.8 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.5 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.1 | 0.5 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.3 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 1.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.3 | GO:1990768 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.1 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 1.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.3 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 0.2 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.1 | 0.3 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.8 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.2 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.1 | 0.3 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.3 | GO:0050992 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.1 | 0.8 | GO:0034127 | regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034127) negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.1 | 0.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.4 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.6 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.3 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.3 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.5 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.1 | 0.2 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 0.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.1 | 0.3 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 1.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.2 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.1 | 0.6 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.1 | 0.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.3 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.5 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.3 | GO:0045861 | negative regulation of proteolysis(GO:0045861) |
| 0.1 | 0.4 | GO:0034091 | regulation of maintenance of sister chromatid cohesion(GO:0034091) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) |
| 0.1 | 0.2 | GO:1903236 | regulation of leukocyte tethering or rolling(GO:1903236) negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.8 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.4 | GO:0002353 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.6 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.4 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.1 | 0.3 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.0 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.5 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.1 | 0.4 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 0.3 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.4 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.2 | GO:0007135 | meiosis II(GO:0007135) |
| 0.1 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.6 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.3 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 1.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.2 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.1 | 0.4 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.1 | 0.5 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.2 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.1 | 0.2 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.3 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.3 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.3 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.0 | 0.1 | GO:0032106 | positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.0 | 0.4 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.3 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.4 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.2 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.4 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 1.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.3 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.4 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.4 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.5 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.3 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0070848 | response to growth factor(GO:0070848) |
| 0.0 | 4.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.5 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.3 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.4 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.3 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.5 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.2 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.2 | GO:2000275 | cellular amide catabolic process(GO:0043605) regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.2 | GO:0010983 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.3 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.3 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) ribonucleoprotein complex export from nucleus(GO:0071426) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0070839 | spleen trabecula formation(GO:0060345) divalent metal ion export(GO:0070839) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.0 | 0.9 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.2 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.6 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.2 | GO:2000109 | regulation of macrophage apoptotic process(GO:2000109) |
| 0.0 | 0.5 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.2 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:0060901 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.0 | 0.3 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 1.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.3 | GO:0046075 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 1.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.3 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.4 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 1.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0071971 | extracellular exosome assembly(GO:0071971) regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.7 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.6 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0090314 | regulation of protein targeting to membrane(GO:0090313) positive regulation of protein targeting to membrane(GO:0090314) |
| 0.0 | 0.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.9 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.3 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.5 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0070428 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.8 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.7 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 1.0 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.6 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.3 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.1 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:1990927 | vesicle-mediated cholesterol transport(GO:0090119) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.1 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.2 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.2 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.8 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:1904237 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:0010796 | regulation of multivesicular body size(GO:0010796) regulation of endosome size(GO:0051036) |
| 0.0 | 0.2 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.1 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 1.4 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 1.2 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.3 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.2 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.1 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 1.1 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.4 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0030856 | regulation of epithelial cell differentiation(GO:0030856) |
| 0.0 | 0.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 1.3 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0061528 | gamma-aminobutyric acid biosynthetic process(GO:0009449) negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.3 | GO:0051095 | regulation of helicase activity(GO:0051095) |
| 0.0 | 0.4 | GO:0050812 | regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.5 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.2 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.3 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:1900920 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 0.0 | 0.7 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 | 0.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.0 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0032446 | protein modification by small protein conjugation(GO:0032446) |
| 0.0 | 0.1 | GO:0045988 | negative regulation of striated muscle contraction(GO:0045988) |
| 0.0 | 0.1 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:2000861 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0039007 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.1 | GO:0060701 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.0 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.2 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.0 | GO:0070873 | regulation of glycogen metabolic process(GO:0070873) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 1.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.0 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.1 | GO:2000410 | regulation of thymocyte migration(GO:2000410) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.0 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.3 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:0060939 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0015820 | leucine transport(GO:0015820) |
| 0.0 | 0.0 | GO:0015827 | tryptophan transport(GO:0015827) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 1.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 1.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.4 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0010875 | positive regulation of cholesterol efflux(GO:0010875) |
| 0.0 | 0.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.1 | GO:0003018 | vascular process in circulatory system(GO:0003018) regulation of tube size(GO:0035150) regulation of blood vessel size(GO:0050880) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.0 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.0 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.1 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.4 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.1 | GO:0071219 | cellular response to biotic stimulus(GO:0071216) cellular response to molecule of bacterial origin(GO:0071219) |
| 0.0 | 0.1 | GO:0022011 | Schwann cell development(GO:0014044) myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.5 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.0 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.2 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.0 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.7 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.4 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.6 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.0 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.0 | 0.1 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.2 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.2 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 0.0 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.0 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.0 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.0 | GO:0044320 | cellular response to leptin stimulus(GO:0044320) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.0 | GO:1904798 | regulation of core promoter binding(GO:1904796) positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.2 | GO:0032401 | establishment of melanosome localization(GO:0032401) |
| 0.0 | 0.0 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.0 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.0 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.2 | 0.6 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.2 | 0.6 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.2 | 0.5 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.2 | 0.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.4 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 0.1 | GO:0097451 | astrocyte end-foot(GO:0097450) glial limiting end-foot(GO:0097451) |
| 0.1 | 0.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 0.4 | GO:0070993 | translation preinitiation complex(GO:0070993) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 1.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.6 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 0.3 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.1 | 0.3 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.3 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.3 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 1.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.2 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.1 | 0.8 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.7 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.8 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.3 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.2 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.1 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.7 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.8 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.1 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.9 | GO:0030426 | growth cone(GO:0030426) site of polarized growth(GO:0030427) |
| 0.1 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.5 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.3 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 1.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.2 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 1.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.0 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.5 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.4 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.5 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.6 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 1.2 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.0 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 1.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.1 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 2.2 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 1.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.0 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 2.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.3 | 0.8 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.2 | 1.0 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.2 | 0.9 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.2 | 2.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 1.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 0.6 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 0.6 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.2 | 0.6 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.2 | 0.7 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.2 | 0.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.2 | 1.0 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.2 | 0.7 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.2 | 0.6 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.2 | 1.2 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.1 | 0.4 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.4 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.4 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 0.4 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.1 | 0.4 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 0.8 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.4 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 0.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 1.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 0.4 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.3 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.1 | 0.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.3 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 0.8 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.4 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.1 | 1.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.3 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.6 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.3 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.5 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 0.7 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.1 | 0.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 1.4 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.9 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.5 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.2 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.1 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.3 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 0.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.2 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.1 | 0.2 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.3 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 0.5 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.3 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.1 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.3 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.1 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.5 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.1 | GO:0004659 | polyprenyltransferase activity(GO:0002094) prenyltransferase activity(GO:0004659) |
| 0.1 | 1.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 1.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.4 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.1 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.3 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 0.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.2 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.6 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.5 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.3 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 1.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.0 | 0.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 2.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.4 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:1902379 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.0 | 0.1 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.0 | 0.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.4 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.2 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.3 | GO:0032405 | MutLalpha complex binding(GO:0032405) MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.3 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.1 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.0 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.3 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.9 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.0 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 2.0 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.0 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.0 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 0.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.6 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.1 | 1.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.9 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.4 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.7 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 1.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 1.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.0 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.4 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |