Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
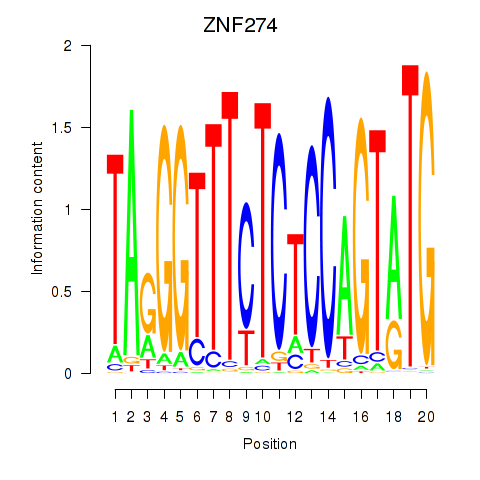
Results for ZNF274
Z-value: 0.49
Transcription factors associated with ZNF274
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF274
|
ENSG00000171606.13 | zinc finger protein 274 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF274 | hg19_v2_chr19_+_58694742_58694863 | 0.45 | 3.7e-01 | Click! |
Activity profile of ZNF274 motif
Sorted Z-values of ZNF274 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_56109119 | 0.74 |
ENST00000587678.1
|
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr11_+_72281681 | 0.35 |
ENST00000450804.3
|
RP11-169D4.1
|
RP11-169D4.1 |
| chr7_+_30791743 | 0.24 |
ENST00000013222.5
ENST00000409539.1 |
INMT
|
indolethylamine N-methyltransferase |
| chr17_-_73636019 | 0.22 |
ENST00000580707.1
|
RECQL5
|
RecQ protein-like 5 |
| chr15_+_89164560 | 0.20 |
ENST00000379231.3
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr5_+_121465234 | 0.20 |
ENST00000504912.1
ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chrX_+_52920336 | 0.18 |
ENST00000452154.2
|
FAM156B
|
family with sequence similarity 156, member B |
| chr19_+_7584088 | 0.16 |
ENST00000394341.2
|
ZNF358
|
zinc finger protein 358 |
| chr10_-_29811456 | 0.16 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr1_-_206671061 | 0.15 |
ENST00000367119.1
|
C1orf147
|
chromosome 1 open reading frame 147 |
| chr20_+_60174827 | 0.14 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr16_+_29819372 | 0.14 |
ENST00000568544.1
ENST00000569978.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr2_+_11864458 | 0.14 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chr9_+_125512019 | 0.13 |
ENST00000373684.1
ENST00000304720.2 |
OR1L6
|
olfactory receptor, family 1, subfamily L, member 6 |
| chr13_-_74993252 | 0.13 |
ENST00000325811.1
|
AL355390.1
|
Uncharacterized protein |
| chr5_+_121465207 | 0.13 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr16_+_29127282 | 0.12 |
ENST00000562902.1
|
RP11-426C22.5
|
RP11-426C22.5 |
| chrX_+_16185604 | 0.12 |
ENST00000400004.2
|
MAGEB17
|
melanoma antigen family B, 17 |
| chr7_+_6713376 | 0.12 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr16_+_29819096 | 0.11 |
ENST00000568411.1
ENST00000563012.1 ENST00000562557.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr19_+_6135646 | 0.10 |
ENST00000588304.1
ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr15_+_89164520 | 0.10 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease |
| chr19_-_19774473 | 0.09 |
ENST00000357324.6
|
ATP13A1
|
ATPase type 13A1 |
| chr6_+_31553901 | 0.09 |
ENST00000418507.2
ENST00000438075.2 ENST00000376100.3 ENST00000376111.4 |
LST1
|
leukocyte specific transcript 1 |
| chr17_+_41132564 | 0.09 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr6_+_30687978 | 0.09 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr16_+_29819446 | 0.08 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr6_+_31553978 | 0.08 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr22_-_43036607 | 0.08 |
ENST00000505920.1
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr6_+_36165133 | 0.07 |
ENST00000446974.1
ENST00000454960.1 |
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr1_-_54355430 | 0.07 |
ENST00000371399.1
ENST00000072644.1 ENST00000412288.1 |
YIPF1
|
Yip1 domain family, member 1 |
| chr17_+_73028676 | 0.07 |
ENST00000581589.1
|
KCTD2
|
potassium channel tetramerization domain containing 2 |
| chr1_+_45205498 | 0.06 |
ENST00000372218.4
|
KIF2C
|
kinesin family member 2C |
| chr3_+_37903432 | 0.06 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr19_+_46498704 | 0.06 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr16_+_31470143 | 0.05 |
ENST00000457010.2
ENST00000563544.1 |
ARMC5
|
armadillo repeat containing 5 |
| chr16_+_31470179 | 0.05 |
ENST00000538189.1
ENST00000268314.4 |
ARMC5
|
armadillo repeat containing 5 |
| chr3_-_38992052 | 0.05 |
ENST00000302328.3
ENST00000450244.1 ENST00000444237.2 |
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr16_+_29818857 | 0.05 |
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr2_-_96971259 | 0.05 |
ENST00000349783.5
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5) |
| chr17_-_41132410 | 0.04 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr3_-_137893721 | 0.04 |
ENST00000505015.2
ENST00000260803.4 |
DBR1
|
debranching RNA lariats 1 |
| chr3_+_50284321 | 0.04 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr20_-_1638360 | 0.04 |
ENST00000216927.4
ENST00000344103.4 |
SIRPG
|
signal-regulatory protein gamma |
| chr22_+_24038593 | 0.03 |
ENST00000452208.1
|
RGL4
|
ral guanine nucleotide dissociation stimulator-like 4 |
| chr2_+_9696028 | 0.03 |
ENST00000607241.1
|
RP11-214N9.1
|
RP11-214N9.1 |
| chr13_+_103459704 | 0.02 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr2_+_159313452 | 0.02 |
ENST00000389757.3
ENST00000389759.3 |
PKP4
|
plakophilin 4 |
| chr2_+_178977143 | 0.02 |
ENST00000286070.5
|
RBM45
|
RNA binding motif protein 45 |
| chr10_+_5488564 | 0.02 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chrX_+_16185836 | 0.01 |
ENST00000400003.1
|
MAGEB17
|
melanoma antigen family B, 17 |
| chr18_+_9885934 | 0.01 |
ENST00000357775.5
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr20_-_1638408 | 0.01 |
ENST00000303415.3
ENST00000381583.2 |
SIRPG
|
signal-regulatory protein gamma |
| chr2_-_42991257 | 0.01 |
ENST00000378661.2
|
OXER1
|
oxoeicosanoid (OXE) receptor 1 |
| chr12_-_49581152 | 0.00 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF274
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |