Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
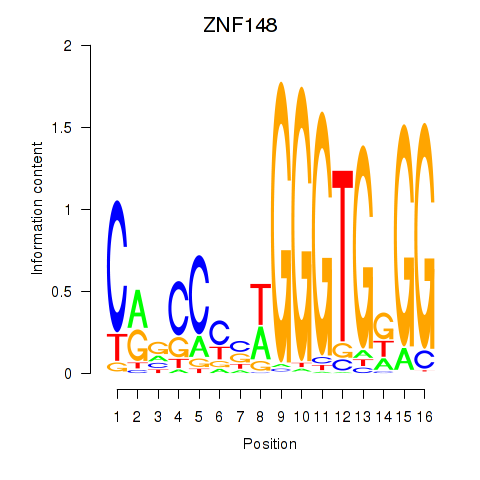
Results for ZNF148
Z-value: 0.91
Transcription factors associated with ZNF148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF148
|
ENSG00000163848.14 | zinc finger protein 148 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF148 | hg19_v2_chr3_-_125094093_125094198 | 0.51 | 3.0e-01 | Click! |
Activity profile of ZNF148 motif
Sorted Z-values of ZNF148 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_10199566 | 0.38 |
ENST00000430336.1
|
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr19_+_10197463 | 0.36 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr3_-_49203744 | 0.36 |
ENST00000321895.6
|
CCDC71
|
coiled-coil domain containing 71 |
| chr11_+_46402297 | 0.36 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr11_+_46402482 | 0.34 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr5_+_172484377 | 0.34 |
ENST00000523161.1
|
CREBRF
|
CREB3 regulatory factor |
| chr17_+_41177220 | 0.33 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr19_-_53662257 | 0.33 |
ENST00000599096.1
ENST00000334197.7 ENST00000597183.1 ENST00000601804.1 ENST00000601469.2 ENST00000452676.2 |
ZNF347
|
zinc finger protein 347 |
| chr10_-_77161004 | 0.32 |
ENST00000418818.2
|
RP11-399K21.11
|
RP11-399K21.11 |
| chr14_-_24733444 | 0.30 |
ENST00000560478.1
ENST00000560443.1 |
TGM1
|
transglutaminase 1 |
| chr9_+_35673853 | 0.30 |
ENST00000378357.4
|
CA9
|
carbonic anhydrase IX |
| chr4_+_75480629 | 0.30 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr9_-_34523027 | 0.29 |
ENST00000399775.2
|
ENHO
|
energy homeostasis associated |
| chr5_-_141030943 | 0.28 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr19_+_18718214 | 0.28 |
ENST00000600490.1
|
TMEM59L
|
transmembrane protein 59-like |
| chr1_-_11714700 | 0.28 |
ENST00000354287.4
|
FBXO2
|
F-box protein 2 |
| chr11_+_46402583 | 0.28 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr6_+_73331520 | 0.28 |
ENST00000342056.2
ENST00000355194.4 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr19_-_12551849 | 0.28 |
ENST00000595562.1
ENST00000301547.5 |
CTD-3105H18.16
ZNF443
|
Uncharacterized protein zinc finger protein 443 |
| chr7_+_149416439 | 0.27 |
ENST00000497895.1
|
KRBA1
|
KRAB-A domain containing 1 |
| chr8_+_144821557 | 0.26 |
ENST00000534398.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr15_-_74494779 | 0.26 |
ENST00000571341.1
|
STRA6
|
stimulated by retinoic acid 6 |
| chr17_-_7197881 | 0.25 |
ENST00000007699.5
|
YBX2
|
Y box binding protein 2 |
| chr19_-_51071302 | 0.25 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr2_-_47572207 | 0.25 |
ENST00000441997.1
|
AC073283.4
|
AC073283.4 |
| chr4_+_75310851 | 0.24 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr13_-_52027134 | 0.24 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr20_+_19738792 | 0.24 |
ENST00000412571.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chr19_-_56048456 | 0.24 |
ENST00000413299.1
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr19_-_8942962 | 0.23 |
ENST00000601372.1
|
ZNF558
|
zinc finger protein 558 |
| chr20_+_10199468 | 0.23 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr3_+_155588375 | 0.23 |
ENST00000295920.7
|
GMPS
|
guanine monphosphate synthase |
| chr12_+_132312931 | 0.23 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr20_-_35724388 | 0.22 |
ENST00000344359.3
ENST00000373664.3 |
RBL1
|
retinoblastoma-like 1 (p107) |
| chr4_+_75311019 | 0.22 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr6_-_31138439 | 0.21 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr12_+_50479109 | 0.21 |
ENST00000550477.1
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr11_-_96123022 | 0.21 |
ENST00000542662.1
|
CCDC82
|
coiled-coil domain containing 82 |
| chr10_-_131640435 | 0.20 |
ENST00000440978.1
|
EBF3
|
early B-cell factor 3 |
| chr1_+_111682827 | 0.20 |
ENST00000357172.4
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr8_+_104383759 | 0.20 |
ENST00000415886.2
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr1_+_27113963 | 0.20 |
ENST00000430292.1
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr2_+_220306745 | 0.19 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr2_-_75745823 | 0.19 |
ENST00000452003.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr15_-_59981479 | 0.19 |
ENST00000607373.1
|
BNIP2
|
BCL2/adenovirus E1B 19kDa interacting protein 2 |
| chr1_-_150208498 | 0.19 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_+_93646238 | 0.18 |
ENST00000448243.1
ENST00000370276.1 |
CCDC18
|
coiled-coil domain containing 18 |
| chr10_-_69835001 | 0.18 |
ENST00000513996.1
ENST00000412272.2 ENST00000395198.3 ENST00000492996.2 |
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr8_+_38758845 | 0.18 |
ENST00000519640.1
|
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr22_-_32341336 | 0.18 |
ENST00000248984.3
|
C22orf24
|
chromosome 22 open reading frame 24 |
| chr10_-_69834973 | 0.17 |
ENST00000395187.2
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr22_+_24115000 | 0.17 |
ENST00000215743.3
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3) |
| chrX_-_39923656 | 0.17 |
ENST00000413905.1
|
BCOR
|
BCL6 corepressor |
| chr6_+_30130969 | 0.17 |
ENST00000376694.4
|
TRIM15
|
tripartite motif containing 15 |
| chr13_-_44361025 | 0.17 |
ENST00000261488.6
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr19_+_10196781 | 0.17 |
ENST00000253110.11
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr3_+_50192457 | 0.16 |
ENST00000414301.1
ENST00000450338.1 |
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chrX_+_49644470 | 0.16 |
ENST00000508866.2
|
USP27X
|
ubiquitin specific peptidase 27, X-linked |
| chr15_+_78833105 | 0.16 |
ENST00000558341.1
ENST00000559437.1 |
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr1_+_225117350 | 0.16 |
ENST00000413949.2
ENST00000430092.1 ENST00000366850.3 ENST00000400952.3 ENST00000366849.1 |
DNAH14
|
dynein, axonemal, heavy chain 14 |
| chr1_+_53308398 | 0.16 |
ENST00000371528.1
|
ZYG11A
|
zyg-11 family member A, cell cycle regulator |
| chr1_-_150208412 | 0.16 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr17_-_40897043 | 0.16 |
ENST00000428826.2
ENST00000592492.1 ENST00000585893.1 ENST00000593214.1 ENST00000590078.1 ENST00000586382.1 ENST00000415827.2 ENST00000592743.1 ENST00000586089.1 ENST00000435174.1 |
EZH1
|
enhancer of zeste homolog 1 (Drosophila) |
| chr4_+_175204865 | 0.16 |
ENST00000505124.1
|
CEP44
|
centrosomal protein 44kDa |
| chr19_+_41860047 | 0.16 |
ENST00000604123.1
|
TMEM91
|
transmembrane protein 91 |
| chr1_+_204485571 | 0.16 |
ENST00000454264.2
ENST00000367183.3 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr1_-_2718286 | 0.15 |
ENST00000401094.6
|
TTC34
|
tetratricopeptide repeat domain 34 |
| chr15_+_78833071 | 0.15 |
ENST00000559365.1
|
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr7_+_73868120 | 0.15 |
ENST00000265755.3
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr1_-_93645818 | 0.15 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr1_-_150208320 | 0.15 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_-_63849571 | 0.15 |
ENST00000295899.5
|
THOC7
|
THO complex 7 homolog (Drosophila) |
| chr22_+_35653445 | 0.15 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr17_-_48207115 | 0.15 |
ENST00000511964.1
|
SAMD14
|
sterile alpha motif domain containing 14 |
| chr14_+_36295638 | 0.15 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr8_-_11873043 | 0.15 |
ENST00000527396.1
|
RP11-481A20.11
|
Protein LOC101060662 |
| chr7_-_140098257 | 0.15 |
ENST00000340308.3
ENST00000447932.2 ENST00000429996.2 ENST00000469193.1 ENST00000326232.9 |
SLC37A3
|
solute carrier family 37, member 3 |
| chr3_-_42845922 | 0.15 |
ENST00000452906.2
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr22_-_30661807 | 0.15 |
ENST00000403389.1
|
OSM
|
oncostatin M |
| chr9_-_100935043 | 0.15 |
ENST00000343933.5
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr19_-_36297632 | 0.15 |
ENST00000588266.2
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr1_-_150208363 | 0.14 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr6_-_32152020 | 0.14 |
ENST00000375055.2
|
AGER
|
advanced glycosylation end product-specific receptor |
| chr2_-_27333965 | 0.14 |
ENST00000452318.2
|
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr15_-_43785274 | 0.14 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr1_-_150208291 | 0.14 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_+_33546714 | 0.14 |
ENST00000294517.6
ENST00000358680.3 ENST00000373443.3 ENST00000398167.1 |
ADC
|
arginine decarboxylase |
| chr1_+_35225339 | 0.14 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr19_+_7599792 | 0.14 |
ENST00000600942.1
ENST00000593924.1 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr3_-_16555150 | 0.14 |
ENST00000334133.4
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr1_-_95007193 | 0.14 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr12_-_95467267 | 0.14 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr19_-_12945362 | 0.14 |
ENST00000590404.1
ENST00000592204.1 |
RTBDN
|
retbindin |
| chr16_+_89238149 | 0.13 |
ENST00000289746.2
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr10_+_70748487 | 0.13 |
ENST00000361983.4
|
KIAA1279
|
KIAA1279 |
| chr12_+_34175398 | 0.13 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chrX_+_152990302 | 0.13 |
ENST00000218104.3
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr4_-_185726906 | 0.13 |
ENST00000513317.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr15_-_76352069 | 0.13 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chrX_-_118739835 | 0.13 |
ENST00000542113.1
ENST00000304449.5 |
NKRF
|
NFKB repressing factor |
| chr11_-_34937858 | 0.13 |
ENST00000278359.5
|
APIP
|
APAF1 interacting protein |
| chr14_-_23791484 | 0.13 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr6_+_149638876 | 0.13 |
ENST00000392282.1
|
TAB2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr6_-_33548006 | 0.13 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr19_+_10196981 | 0.12 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr8_+_48920960 | 0.12 |
ENST00000523111.2
ENST00000523432.1 ENST00000521346.1 ENST00000517630.1 |
UBE2V2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr3_+_49977440 | 0.12 |
ENST00000442092.1
ENST00000266022.4 ENST00000443081.1 |
RBM6
|
RNA binding motif protein 6 |
| chr3_-_88108192 | 0.12 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr12_+_110940005 | 0.12 |
ENST00000409246.1
ENST00000392672.4 ENST00000409300.1 ENST00000409425.1 |
RAD9B
|
RAD9 homolog B (S. pombe) |
| chr1_-_166136187 | 0.12 |
ENST00000338353.3
|
FAM78B
|
family with sequence similarity 78, member B |
| chr1_-_149889382 | 0.12 |
ENST00000369145.1
ENST00000369146.3 |
SV2A
|
synaptic vesicle glycoprotein 2A |
| chr6_-_33547975 | 0.12 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr6_+_89790459 | 0.12 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr7_+_73868220 | 0.12 |
ENST00000455841.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr6_+_31633833 | 0.12 |
ENST00000375882.2
ENST00000375880.2 |
CSNK2B
CSNK2B-LY6G5B-1181
|
casein kinase 2, beta polypeptide Uncharacterized protein |
| chr1_+_242011468 | 0.12 |
ENST00000366548.3
|
EXO1
|
exonuclease 1 |
| chr2_-_47168850 | 0.12 |
ENST00000409207.1
|
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr12_-_90102594 | 0.12 |
ENST00000428670.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr17_-_46507567 | 0.12 |
ENST00000584924.1
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr2_-_47168906 | 0.12 |
ENST00000444761.2
ENST00000409147.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr14_+_31343951 | 0.12 |
ENST00000556908.1
ENST00000555881.1 ENST00000460581.2 |
COCH
|
cochlin |
| chr14_+_31494841 | 0.12 |
ENST00000556232.1
ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr9_-_100954910 | 0.12 |
ENST00000375077.4
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr5_+_138678131 | 0.12 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr1_+_242011485 | 0.12 |
ENST00000423131.1
ENST00000523590.1 |
EXO1
|
exonuclease 1 |
| chr19_-_51522955 | 0.11 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr1_-_92351666 | 0.11 |
ENST00000465892.2
ENST00000417833.2 |
TGFBR3
|
transforming growth factor, beta receptor III |
| chr5_-_176900610 | 0.11 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr14_+_31343747 | 0.11 |
ENST00000216361.4
ENST00000396618.3 ENST00000475087.1 |
COCH
|
cochlin |
| chr11_-_34938039 | 0.11 |
ENST00000395787.3
|
APIP
|
APAF1 interacting protein |
| chr20_-_47894569 | 0.11 |
ENST00000371744.1
ENST00000371752.1 ENST00000396105.1 |
ZNFX1
|
zinc finger, NFX1-type containing 1 |
| chr7_-_80548667 | 0.11 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr2_+_28974489 | 0.11 |
ENST00000455580.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr17_+_39968926 | 0.11 |
ENST00000585664.1
ENST00000585922.1 ENST00000429461.1 |
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr11_+_34938119 | 0.11 |
ENST00000227868.4
ENST00000430469.2 ENST00000533262.1 |
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr10_+_134000404 | 0.11 |
ENST00000338492.4
ENST00000368629.1 |
DPYSL4
|
dihydropyrimidinase-like 4 |
| chr20_-_31124186 | 0.11 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr12_+_56137064 | 0.11 |
ENST00000257868.5
ENST00000546799.1 |
GDF11
|
growth differentiation factor 11 |
| chr2_+_220408724 | 0.11 |
ENST00000421791.1
ENST00000373883.3 ENST00000451952.1 |
TMEM198
|
transmembrane protein 198 |
| chr15_+_79165372 | 0.11 |
ENST00000558502.1
|
MORF4L1
|
mortality factor 4 like 1 |
| chr15_+_40544749 | 0.11 |
ENST00000559617.1
ENST00000560684.1 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr1_-_40367668 | 0.11 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr1_-_155224699 | 0.11 |
ENST00000491082.1
|
FAM189B
|
family with sequence similarity 189, member B |
| chr17_+_40950797 | 0.11 |
ENST00000588408.1
ENST00000585355.1 |
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr17_-_40540586 | 0.11 |
ENST00000264657.5
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr12_+_50479101 | 0.11 |
ENST00000551966.1
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr15_-_74044719 | 0.10 |
ENST00000559817.1
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr9_+_110046334 | 0.10 |
ENST00000416373.2
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr17_+_7348658 | 0.10 |
ENST00000570557.1
ENST00000536404.2 ENST00000576360.1 |
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr3_-_197024965 | 0.10 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr19_+_48958766 | 0.10 |
ENST00000342291.2
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chrX_+_73641286 | 0.10 |
ENST00000587091.1
|
SLC16A2
|
solute carrier family 16, member 2 (thyroid hormone transporter) |
| chr5_-_41510725 | 0.10 |
ENST00000328457.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr19_-_45579762 | 0.10 |
ENST00000303809.2
|
ZNF296
|
zinc finger protein 296 |
| chr14_+_31494672 | 0.10 |
ENST00000542754.2
ENST00000313566.6 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr1_+_183155373 | 0.10 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr1_-_98386543 | 0.10 |
ENST00000423006.2
ENST00000370192.3 ENST00000306031.5 |
DPYD
|
dihydropyrimidine dehydrogenase |
| chr19_-_36297348 | 0.10 |
ENST00000589835.1
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr5_+_95998673 | 0.10 |
ENST00000514845.1
|
CAST
|
calpastatin |
| chr19_-_663171 | 0.10 |
ENST00000606896.1
ENST00000589762.2 |
RNF126
|
ring finger protein 126 |
| chr8_-_25315905 | 0.10 |
ENST00000221200.4
|
KCTD9
|
potassium channel tetramerization domain containing 9 |
| chr10_+_22610876 | 0.09 |
ENST00000442508.1
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr12_-_95467356 | 0.09 |
ENST00000393101.3
ENST00000333003.5 |
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr9_-_35618364 | 0.09 |
ENST00000378431.1
ENST00000378430.3 ENST00000259633.4 |
CD72
|
CD72 molecule |
| chr4_+_52709229 | 0.09 |
ENST00000334635.5
ENST00000381441.3 ENST00000381437.4 |
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 |
| chr7_+_144052381 | 0.09 |
ENST00000498580.1
ENST00000056217.5 |
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr10_-_32217717 | 0.09 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr19_-_19626351 | 0.09 |
ENST00000585580.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr15_+_78556809 | 0.09 |
ENST00000343789.3
ENST00000394852.3 |
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr2_+_37571717 | 0.09 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr11_-_118661828 | 0.09 |
ENST00000264018.4
|
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr1_-_63782888 | 0.09 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr12_-_124457163 | 0.09 |
ENST00000535556.1
|
CCDC92
|
coiled-coil domain containing 92 |
| chr18_+_43684310 | 0.09 |
ENST00000592471.1
ENST00000585518.1 |
HAUS1
|
HAUS augmin-like complex, subunit 1 |
| chr11_+_63448955 | 0.09 |
ENST00000377819.5
ENST00000339997.4 ENST00000540798.1 ENST00000545432.1 ENST00000543552.1 ENST00000537981.1 |
RTN3
|
reticulon 3 |
| chr7_-_5465045 | 0.09 |
ENST00000399434.2
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr10_+_94608245 | 0.09 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr1_-_59165763 | 0.09 |
ENST00000472487.1
|
MYSM1
|
Myb-like, SWIRM and MPN domains 1 |
| chr20_-_36793663 | 0.09 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr5_+_76114758 | 0.09 |
ENST00000514165.1
ENST00000296677.4 |
F2RL1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr11_+_64107663 | 0.09 |
ENST00000356786.5
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr2_-_61765315 | 0.09 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr15_+_79165296 | 0.09 |
ENST00000558746.1
ENST00000558830.1 ENST00000559345.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr14_-_23284675 | 0.09 |
ENST00000555959.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr12_+_90102729 | 0.09 |
ENST00000605386.1
|
LINC00936
|
long intergenic non-protein coding RNA 936 |
| chr14_-_88200641 | 0.09 |
ENST00000556168.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr11_-_77184739 | 0.09 |
ENST00000524847.1
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr2_-_38604398 | 0.09 |
ENST00000443098.1
ENST00000449130.1 ENST00000378954.4 ENST00000539122.1 ENST00000419554.2 ENST00000451483.1 ENST00000406122.1 |
ATL2
|
atlastin GTPase 2 |
| chr3_+_50192833 | 0.09 |
ENST00000426511.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr19_-_10464570 | 0.09 |
ENST00000529739.1
|
TYK2
|
tyrosine kinase 2 |
| chr1_+_11714914 | 0.08 |
ENST00000425796.1
ENST00000376770.1 ENST00000376768.1 ENST00000251547.5 ENST00000376760.1 ENST00000376762.4 |
FBXO44
|
F-box protein 44 |
| chr7_+_30960915 | 0.08 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr10_+_5932174 | 0.08 |
ENST00000362091.4
|
FBXO18
|
F-box protein, helicase, 18 |
| chr11_+_66742742 | 0.08 |
ENST00000308963.4
|
C11orf86
|
chromosome 11 open reading frame 86 |
| chr19_+_55587266 | 0.08 |
ENST00000201647.6
ENST00000540810.1 |
EPS8L1
|
EPS8-like 1 |
| chr3_+_49977490 | 0.08 |
ENST00000539992.1
|
RBM6
|
RNA binding motif protein 6 |
| chr19_+_52800410 | 0.08 |
ENST00000595962.1
ENST00000598016.1 ENST00000334564.7 ENST00000490272.1 |
ZNF480
|
zinc finger protein 480 |
| chr17_-_61850894 | 0.08 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr11_-_118661588 | 0.08 |
ENST00000534980.1
ENST00000526070.2 |
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr2_+_153191706 | 0.08 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF148
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 0.6 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.2 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.2 | GO:1904597 | regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904596) negative regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904597) regulation of advanced glycation end-product receptor activity(GO:1904603) negative regulation of advanced glycation end-product receptor activity(GO:1904604) negative regulation of connective tissue replacement(GO:1905204) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.3 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.5 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.2 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.3 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.3 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0019482 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.0 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:1902568 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.0 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901205) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.1 | GO:0060979 | transforming growth factor beta receptor complex assembly(GO:0007181) response to luteinizing hormone(GO:0034699) vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.3 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.0 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.0 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.1 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.8 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.0 | 0.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.0 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.1 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.1 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051908 | double-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0051908) |
| 0.1 | 0.2 | GO:1904599 | advanced glycation end-product binding(GO:1904599) |
| 0.1 | 0.2 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.1 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.2 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.8 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |