Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
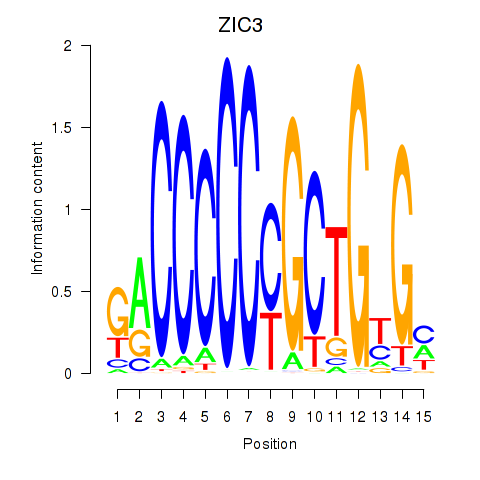
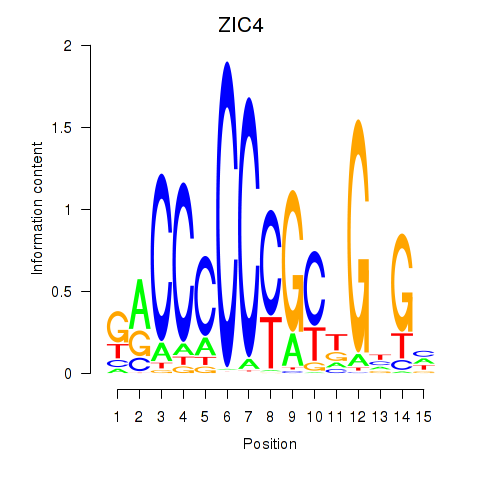
Results for ZIC3_ZIC4
Z-value: 0.38
Transcription factors associated with ZIC3_ZIC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC3
|
ENSG00000156925.7 | Zic family member 3 |
|
ZIC4
|
ENSG00000174963.13 | Zic family member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZIC3 | hg19_v2_chrX_+_136648297_136648319 | 0.38 | 4.6e-01 | Click! |
Activity profile of ZIC3_ZIC4 motif
Sorted Z-values of ZIC3_ZIC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_302918 | 0.25 |
ENST00000599994.1
|
AC187652.1
|
Protein LOC100996433 |
| chr9_-_34662651 | 0.22 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr11_+_73675873 | 0.18 |
ENST00000537753.1
ENST00000542350.1 |
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr17_+_39405939 | 0.18 |
ENST00000334109.2
|
KRTAP9-4
|
keratin associated protein 9-4 |
| chr1_+_154378049 | 0.17 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr2_-_67442409 | 0.15 |
ENST00000414404.1
ENST00000433133.1 |
AC078941.1
|
AC078941.1 |
| chr10_-_9801179 | 0.15 |
ENST00000419836.1
|
RP5-1051H14.2
|
RP5-1051H14.2 |
| chr19_+_10531150 | 0.14 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr9_+_139738867 | 0.13 |
ENST00000436881.1
|
C9orf172
|
chromosome 9 open reading frame 172 |
| chr3_-_118753792 | 0.13 |
ENST00000480431.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr16_+_4364762 | 0.12 |
ENST00000262366.3
|
GLIS2
|
GLIS family zinc finger 2 |
| chr17_-_73840614 | 0.12 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr22_-_41842781 | 0.12 |
ENST00000434408.1
|
TOB2
|
transducer of ERBB2, 2 |
| chr19_-_40790993 | 0.12 |
ENST00000596634.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr11_+_73676281 | 0.11 |
ENST00000543947.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr8_+_1711918 | 0.11 |
ENST00000331222.4
|
CLN8
|
ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) |
| chr19_-_12992244 | 0.11 |
ENST00000538460.1
|
DNASE2
|
deoxyribonuclease II, lysosomal |
| chr3_-_87039662 | 0.11 |
ENST00000494229.1
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr11_-_64014379 | 0.11 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr16_+_2570340 | 0.11 |
ENST00000568263.1
ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2
|
amidohydrolase domain containing 2 |
| chr16_+_2059872 | 0.11 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr1_-_16400086 | 0.11 |
ENST00000375662.4
|
FAM131C
|
family with sequence similarity 131, member C |
| chr20_-_31172598 | 0.11 |
ENST00000201961.2
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr3_+_195413160 | 0.10 |
ENST00000599448.1
|
LINC00969
|
long intergenic non-protein coding RNA 969 |
| chr5_+_72509751 | 0.10 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr19_-_40790729 | 0.10 |
ENST00000423127.1
ENST00000583859.1 ENST00000456441.1 ENST00000452077.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr16_+_28996572 | 0.09 |
ENST00000360872.5
ENST00000566177.1 ENST00000354453.4 |
LAT
|
linker for activation of T cells |
| chr17_-_73840415 | 0.09 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr22_+_29702572 | 0.09 |
ENST00000407647.2
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr11_+_58938903 | 0.09 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr9_-_130742792 | 0.09 |
ENST00000373095.1
|
FAM102A
|
family with sequence similarity 102, member A |
| chr11_-_67272794 | 0.09 |
ENST00000436757.2
ENST00000356404.3 |
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr17_-_78194124 | 0.09 |
ENST00000570427.1
ENST00000570923.1 |
SGSH
|
N-sulfoglucosamine sulfohydrolase |
| chr8_-_145980808 | 0.08 |
ENST00000525191.1
|
ZNF251
|
zinc finger protein 251 |
| chr1_+_3370990 | 0.08 |
ENST00000378378.4
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr19_-_40791302 | 0.08 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr6_-_85473073 | 0.08 |
ENST00000606621.1
|
TBX18
|
T-box 18 |
| chr7_-_135412925 | 0.08 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr17_-_4890919 | 0.08 |
ENST00000572543.1
ENST00000381311.5 ENST00000348066.3 ENST00000358183.4 |
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr17_-_73840774 | 0.08 |
ENST00000207549.4
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chrX_+_71996972 | 0.08 |
ENST00000334036.5
|
DMRTC1B
|
DMRT-like family C1B |
| chr11_-_62477313 | 0.08 |
ENST00000464544.1
ENST00000530009.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr16_+_29466426 | 0.07 |
ENST00000567248.1
|
SLX1B
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr17_-_78194147 | 0.07 |
ENST00000534910.1
ENST00000326317.6 |
SGSH
|
N-sulfoglucosamine sulfohydrolase |
| chr7_-_44924939 | 0.07 |
ENST00000395699.2
|
PURB
|
purine-rich element binding protein B |
| chr11_+_637246 | 0.07 |
ENST00000176183.5
|
DRD4
|
dopamine receptor D4 |
| chr22_-_19466643 | 0.07 |
ENST00000474226.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr1_+_38273988 | 0.07 |
ENST00000446260.2
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr16_+_777739 | 0.07 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr16_-_2059797 | 0.07 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr2_-_107502456 | 0.07 |
ENST00000419159.2
|
ST6GAL2
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 |
| chr9_-_114245938 | 0.07 |
ENST00000602447.1
|
KIAA0368
|
KIAA0368 |
| chr1_-_146644036 | 0.07 |
ENST00000425272.2
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr7_+_150758304 | 0.07 |
ENST00000482950.1
ENST00000463414.1 ENST00000310317.5 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr19_-_40791211 | 0.07 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr19_-_12992274 | 0.07 |
ENST00000592506.1
ENST00000222219.3 |
DNASE2
|
deoxyribonuclease II, lysosomal |
| chr12_+_49932886 | 0.07 |
ENST00000257981.6
|
KCNH3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr9_-_114246332 | 0.07 |
ENST00000602978.1
|
KIAA0368
|
KIAA0368 |
| chr19_-_40724246 | 0.07 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr19_-_40791160 | 0.07 |
ENST00000358335.5
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr1_+_38273818 | 0.07 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr4_+_52917451 | 0.07 |
ENST00000295213.4
ENST00000419395.2 |
SPATA18
|
spermatogenesis associated 18 |
| chr22_-_22221900 | 0.07 |
ENST00000215832.6
ENST00000398822.3 |
MAPK1
|
mitogen-activated protein kinase 1 |
| chr9_+_134269439 | 0.07 |
ENST00000405995.1
|
PRRC2B
|
proline-rich coiled-coil 2B |
| chr15_+_73976715 | 0.07 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr20_+_816695 | 0.07 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr9_+_131580734 | 0.07 |
ENST00000372642.4
|
ENDOG
|
endonuclease G |
| chr19_-_37096139 | 0.06 |
ENST00000585983.1
ENST00000585960.1 ENST00000586115.1 |
ZNF529
|
zinc finger protein 529 |
| chr16_+_58498177 | 0.06 |
ENST00000567454.1
|
NDRG4
|
NDRG family member 4 |
| chr7_+_65540853 | 0.06 |
ENST00000380839.4
ENST00000395332.3 ENST00000362000.5 ENST00000395331.3 |
ASL
|
argininosuccinate lyase |
| chr10_+_71561630 | 0.06 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr17_-_43045439 | 0.06 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chr15_+_31619013 | 0.06 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr19_+_49617581 | 0.06 |
ENST00000391864.3
|
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr7_-_74867509 | 0.06 |
ENST00000426327.3
|
GATSL2
|
GATS protein-like 2 |
| chr5_+_139055055 | 0.06 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr19_+_13906250 | 0.06 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr17_-_78194716 | 0.06 |
ENST00000576707.1
|
SGSH
|
N-sulfoglucosamine sulfohydrolase |
| chr17_+_5981270 | 0.06 |
ENST00000571973.1
|
WSCD1
|
WSC domain containing 1 |
| chr10_-_134145321 | 0.06 |
ENST00000368625.4
ENST00000368619.3 ENST00000456004.1 ENST00000368620.2 |
STK32C
|
serine/threonine kinase 32C |
| chr17_+_45331184 | 0.06 |
ENST00000559488.1
ENST00000571680.1 ENST00000435993.2 |
ITGB3
|
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
| chr17_-_40829026 | 0.06 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr7_+_99775520 | 0.06 |
ENST00000317296.5
ENST00000422690.1 ENST00000439782.1 |
STAG3
|
stromal antigen 3 |
| chr15_-_91565770 | 0.06 |
ENST00000535906.1
ENST00000333371.3 |
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast) |
| chr19_-_49314269 | 0.05 |
ENST00000545387.2
ENST00000316273.6 ENST00000402551.1 ENST00000598162.1 ENST00000599246.1 |
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr5_+_145316120 | 0.05 |
ENST00000359120.4
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr1_-_45308616 | 0.05 |
ENST00000447098.2
ENST00000372192.3 |
PTCH2
|
patched 2 |
| chr11_+_76092353 | 0.05 |
ENST00000530460.1
ENST00000321844.4 |
RP11-111M22.2
|
Homo sapiens putative uncharacterized protein FLJ37770-like (LOC100506127), mRNA. |
| chr5_+_140718396 | 0.05 |
ENST00000394576.2
|
PCDHGA2
|
protocadherin gamma subfamily A, 2 |
| chr1_+_43148059 | 0.05 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr22_-_41636929 | 0.05 |
ENST00000216241.9
|
CHADL
|
chondroadherin-like |
| chr15_-_91565743 | 0.05 |
ENST00000535843.1
|
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast) |
| chr11_-_1036706 | 0.05 |
ENST00000421673.2
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming |
| chr17_-_7531121 | 0.05 |
ENST00000573566.1
ENST00000269298.5 |
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2 |
| chr20_-_56286479 | 0.05 |
ENST00000265626.4
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr16_+_4897912 | 0.05 |
ENST00000545171.1
|
UBN1
|
ubinuclein 1 |
| chr1_-_31381528 | 0.05 |
ENST00000339394.6
|
SDC3
|
syndecan 3 |
| chr5_+_139055021 | 0.05 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr9_-_140082983 | 0.05 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr19_-_39390212 | 0.05 |
ENST00000437828.1
|
SIRT2
|
sirtuin 2 |
| chr19_+_54926621 | 0.05 |
ENST00000376530.3
ENST00000445095.1 ENST00000391739.3 ENST00000376531.3 |
TTYH1
|
tweety family member 1 |
| chr12_-_125052010 | 0.05 |
ENST00000458234.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr18_+_2846972 | 0.05 |
ENST00000254528.3
|
EMILIN2
|
elastin microfibril interfacer 2 |
| chr9_+_115913222 | 0.05 |
ENST00000259392.3
|
SLC31A2
|
solute carrier family 31 (copper transporter), member 2 |
| chr16_+_30205225 | 0.05 |
ENST00000345535.4
ENST00000251303.6 |
SLX1A
|
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae) |
| chr15_+_75074410 | 0.05 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr15_+_75074385 | 0.05 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase |
| chr1_-_151119087 | 0.05 |
ENST00000341697.3
ENST00000368914.3 |
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr10_-_131762105 | 0.05 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr12_+_121163538 | 0.05 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr9_+_116638630 | 0.05 |
ENST00000452710.1
ENST00000374124.4 |
ZNF618
|
zinc finger protein 618 |
| chr11_-_65667997 | 0.05 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr12_-_59314246 | 0.05 |
ENST00000320743.3
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr6_+_15249128 | 0.05 |
ENST00000397311.3
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr20_+_60698180 | 0.05 |
ENST00000361670.3
|
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae) |
| chr1_-_21503337 | 0.05 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr6_-_38607673 | 0.05 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr17_-_1083078 | 0.05 |
ENST00000574266.1
ENST00000302538.5 |
ABR
|
active BCR-related |
| chr17_-_56065540 | 0.05 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr16_+_29465822 | 0.04 |
ENST00000330181.5
ENST00000351581.4 |
SLX1B
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr20_+_35201857 | 0.04 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr2_-_99552620 | 0.04 |
ENST00000428096.1
ENST00000397899.2 ENST00000420294.1 |
KIAA1211L
|
KIAA1211-like |
| chr9_+_140083099 | 0.04 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr1_+_6052700 | 0.04 |
ENST00000378092.1
ENST00000445501.1 |
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr7_+_149571045 | 0.04 |
ENST00000479613.1
ENST00000606024.1 ENST00000464662.1 ENST00000425642.2 |
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2 |
| chr17_+_47572647 | 0.04 |
ENST00000172229.3
|
NGFR
|
nerve growth factor receptor |
| chr11_-_116968987 | 0.04 |
ENST00000434315.2
ENST00000292055.4 ENST00000375288.1 ENST00000542607.1 ENST00000445177.1 ENST00000375300.1 ENST00000446921.2 |
SIK3
|
SIK family kinase 3 |
| chr20_+_35201993 | 0.04 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chrX_-_30326445 | 0.04 |
ENST00000378963.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr19_-_12833361 | 0.04 |
ENST00000592287.1
|
TNPO2
|
transportin 2 |
| chr19_+_984313 | 0.04 |
ENST00000251289.5
ENST00000587001.2 ENST00000607440.1 |
WDR18
|
WD repeat domain 18 |
| chr17_+_59477233 | 0.04 |
ENST00000240328.3
|
TBX2
|
T-box 2 |
| chr7_-_4923259 | 0.04 |
ENST00000536091.1
|
RADIL
|
Ras association and DIL domains |
| chr2_-_109605663 | 0.04 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr1_-_159915386 | 0.04 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr16_-_69364467 | 0.04 |
ENST00000288022.1
|
PDF
|
peptide deformylase (mitochondrial) |
| chr14_+_105886150 | 0.04 |
ENST00000331320.7
ENST00000406191.1 |
MTA1
|
metastasis associated 1 |
| chr16_+_28996364 | 0.04 |
ENST00000564277.1
|
LAT
|
linker for activation of T cells |
| chr7_+_65540780 | 0.04 |
ENST00000304874.9
|
ASL
|
argininosuccinate lyase |
| chr19_+_59055814 | 0.04 |
ENST00000594806.1
ENST00000253024.5 ENST00000341753.6 |
TRIM28
|
tripartite motif containing 28 |
| chr5_+_176731572 | 0.04 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr2_-_98612379 | 0.04 |
ENST00000425805.2
|
TMEM131
|
transmembrane protein 131 |
| chr3_-_112356944 | 0.04 |
ENST00000461431.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr11_+_66790816 | 0.04 |
ENST00000527043.1
|
SYT12
|
synaptotagmin XII |
| chr8_-_135708787 | 0.04 |
ENST00000520356.1
ENST00000520727.1 ENST00000520214.1 ENST00000518191.1 ENST00000429442.2 |
ZFAT
|
zinc finger and AT hook domain containing |
| chr10_+_134145614 | 0.04 |
ENST00000368615.3
ENST00000392638.2 ENST00000344079.5 ENST00000356571.4 ENST00000368614.3 |
LRRC27
|
leucine rich repeat containing 27 |
| chr16_-_88717482 | 0.04 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr5_+_137688285 | 0.04 |
ENST00000314358.5
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr20_+_44563267 | 0.04 |
ENST00000372409.3
|
PCIF1
|
PDX1 C-terminal inhibiting factor 1 |
| chr17_-_21454898 | 0.04 |
ENST00000391411.5
ENST00000412778.3 |
C17orf51
|
chromosome 17 open reading frame 51 |
| chr18_+_13217965 | 0.04 |
ENST00000587905.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr19_-_18385221 | 0.04 |
ENST00000595654.2
ENST00000593659.1 ENST00000599528.1 |
KIAA1683
|
KIAA1683 |
| chr14_-_100841794 | 0.04 |
ENST00000556295.1
ENST00000554820.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr9_+_116638562 | 0.04 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr17_-_4871085 | 0.04 |
ENST00000575142.1
ENST00000206020.3 |
SPAG7
|
sperm associated antigen 7 |
| chr17_-_79623597 | 0.04 |
ENST00000574024.1
ENST00000331056.5 |
PDE6G
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr2_-_98280383 | 0.04 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr16_+_67840986 | 0.04 |
ENST00000561639.1
ENST00000567852.1 ENST00000565148.1 ENST00000388833.3 ENST00000561654.1 ENST00000431934.2 |
TSNAXIP1
|
translin-associated factor X interacting protein 1 |
| chr1_+_85527987 | 0.04 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr2_-_73869508 | 0.04 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr2_+_11273179 | 0.04 |
ENST00000381585.3
ENST00000405022.3 |
C2orf50
|
chromosome 2 open reading frame 50 |
| chr19_+_34287751 | 0.04 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr7_-_752577 | 0.04 |
ENST00000544935.1
ENST00000430040.1 ENST00000456696.2 ENST00000406797.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr4_-_860950 | 0.04 |
ENST00000511980.1
ENST00000510799.1 |
GAK
|
cyclin G associated kinase |
| chr16_+_89753070 | 0.04 |
ENST00000353379.7
ENST00000505473.1 ENST00000564192.1 |
CDK10
|
cyclin-dependent kinase 10 |
| chr5_-_126366500 | 0.04 |
ENST00000308660.5
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
| chr7_-_752074 | 0.04 |
ENST00000360274.4
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr12_-_51785393 | 0.04 |
ENST00000605138.1
ENST00000604381.1 ENST00000605055.1 ENST00000605617.1 |
GALNT6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| chr11_-_65667884 | 0.04 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr19_-_49314169 | 0.04 |
ENST00000597011.1
ENST00000601681.1 |
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr16_-_31085514 | 0.04 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr22_-_19165917 | 0.04 |
ENST00000451283.1
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr17_+_75276643 | 0.04 |
ENST00000589070.1
|
SEPT9
|
septin 9 |
| chr4_+_492985 | 0.04 |
ENST00000296306.7
ENST00000536264.1 ENST00000310340.5 ENST00000453061.2 ENST00000504346.1 ENST00000503111.1 ENST00000383028.4 ENST00000509768.1 |
PIGG
|
phosphatidylinositol glycan anchor biosynthesis, class G |
| chr6_+_33168637 | 0.04 |
ENST00000374677.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr18_+_11752040 | 0.04 |
ENST00000423027.3
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr14_-_100841930 | 0.04 |
ENST00000555031.1
ENST00000553395.1 ENST00000553545.1 ENST00000344102.5 ENST00000556338.1 ENST00000392882.2 ENST00000553934.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr16_+_67465016 | 0.04 |
ENST00000326152.5
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr11_-_57004658 | 0.04 |
ENST00000606794.1
|
APLNR
|
apelin receptor |
| chr15_-_70388599 | 0.04 |
ENST00000560996.1
ENST00000558201.1 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr11_+_120081475 | 0.04 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr17_+_27055798 | 0.04 |
ENST00000268766.6
|
NEK8
|
NIMA-related kinase 8 |
| chr20_+_3870024 | 0.04 |
ENST00000610179.1
|
PANK2
|
pantothenate kinase 2 |
| chr18_+_46065483 | 0.04 |
ENST00000382998.4
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr19_+_33685490 | 0.03 |
ENST00000253193.7
|
LRP3
|
low density lipoprotein receptor-related protein 3 |
| chr16_-_30204987 | 0.03 |
ENST00000569282.1
ENST00000567436.1 |
BOLA2B
|
bolA family member 2B |
| chr20_-_30458432 | 0.03 |
ENST00000375966.4
ENST00000278979.3 |
DUSP15
|
dual specificity phosphatase 15 |
| chr16_+_28834531 | 0.03 |
ENST00000570200.1
|
ATXN2L
|
ataxin 2-like |
| chr9_-_139371533 | 0.03 |
ENST00000290037.6
ENST00000431893.2 ENST00000371706.3 |
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr7_-_99774945 | 0.03 |
ENST00000292377.2
|
GPC2
|
glypican 2 |
| chr17_-_40828969 | 0.03 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr11_+_47291193 | 0.03 |
ENST00000428807.1
ENST00000402799.1 ENST00000406482.1 ENST00000349238.3 ENST00000311027.5 ENST00000407859.3 ENST00000395344.3 ENST00000444117.1 |
MADD
|
MAP-kinase activating death domain |
| chr16_-_18470696 | 0.03 |
ENST00000427999.2
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr17_+_38219063 | 0.03 |
ENST00000584985.1
ENST00000264637.4 ENST00000450525.2 |
THRA
|
thyroid hormone receptor, alpha |
| chr6_+_33168597 | 0.03 |
ENST00000374675.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr7_-_5569588 | 0.03 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chr11_-_32457075 | 0.03 |
ENST00000448076.3
|
WT1
|
Wilms tumor 1 |
| chr1_+_201617450 | 0.03 |
ENST00000295624.6
ENST00000367297.4 ENST00000367300.3 |
NAV1
|
neuron navigator 1 |
| chr22_-_47134077 | 0.03 |
ENST00000541677.1
ENST00000216264.8 |
CERK
|
ceramide kinase |
| chr19_+_48949030 | 0.03 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC3_ZIC4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 0.4 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.0 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.1 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.1 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0051586 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.0 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.1 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.0 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016250 | N-sulfoglucosamine sulfohydrolase activity(GO:0016250) hydrolase activity, acting on acid sulfur-nitrogen bonds(GO:0016826) |
| 0.1 | 0.2 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.1 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.2 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.0 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |