Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
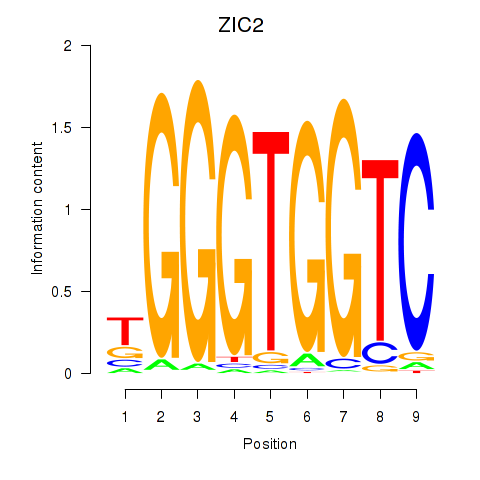
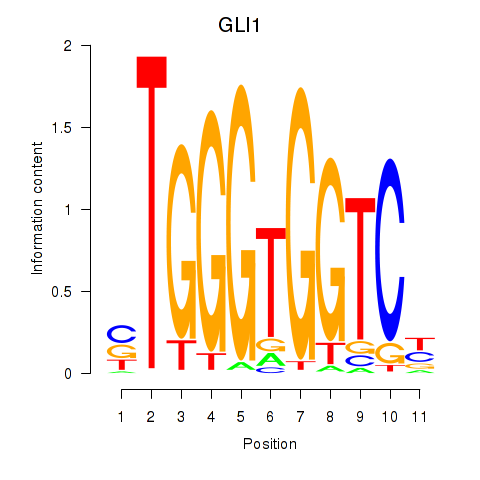
Results for ZIC2_GLI1
Z-value: 0.62
Transcription factors associated with ZIC2_GLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC2
|
ENSG00000043355.6 | Zic family member 2 |
|
GLI1
|
ENSG00000111087.5 | GLI family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI1 | hg19_v2_chr12_+_57854274_57854274 | 0.59 | 2.2e-01 | Click! |
| ZIC2 | hg19_v2_chr13_+_100634004_100634026 | 0.06 | 9.1e-01 | Click! |
Activity profile of ZIC2_GLI1 motif
Sorted Z-values of ZIC2_GLI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_40835076 | 0.55 |
ENST00000591765.1
|
CCR10
|
chemokine (C-C motif) receptor 10 |
| chr14_+_24584056 | 0.36 |
ENST00000561001.1
|
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr18_-_44775554 | 0.32 |
ENST00000425639.1
ENST00000400404.1 |
SKOR2
|
SKI family transcriptional corepressor 2 |
| chr17_-_37558776 | 0.31 |
ENST00000577399.1
|
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr6_-_29600559 | 0.30 |
ENST00000476670.1
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr1_+_36621174 | 0.27 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr12_-_6798025 | 0.24 |
ENST00000542351.1
ENST00000538829.1 |
ZNF384
|
zinc finger protein 384 |
| chr1_+_36621529 | 0.22 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr17_+_43238438 | 0.22 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr16_-_21452040 | 0.20 |
ENST00000521589.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr5_-_138739739 | 0.20 |
ENST00000514983.1
ENST00000507779.2 ENST00000451821.2 ENST00000450845.2 ENST00000509959.1 ENST00000302091.5 |
SPATA24
|
spermatogenesis associated 24 |
| chr8_+_94752349 | 0.20 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr12_-_57030096 | 0.20 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr1_+_209941942 | 0.20 |
ENST00000487271.1
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr17_+_76422409 | 0.19 |
ENST00000600087.1
|
AC061992.1
|
Uncharacterized protein |
| chr22_-_46373004 | 0.19 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr10_-_95360983 | 0.18 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr1_+_226736446 | 0.18 |
ENST00000366788.3
ENST00000366789.4 |
C1orf95
|
chromosome 1 open reading frame 95 |
| chr17_+_40834580 | 0.18 |
ENST00000264638.4
|
CNTNAP1
|
contactin associated protein 1 |
| chr11_+_45168182 | 0.17 |
ENST00000526442.1
|
PRDM11
|
PR domain containing 11 |
| chr17_-_7137582 | 0.16 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr19_-_44285401 | 0.16 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr16_+_86612112 | 0.16 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr20_-_62493217 | 0.15 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135 |
| chr12_-_6798616 | 0.14 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chrX_+_51928002 | 0.14 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chr11_-_65321198 | 0.13 |
ENST00000530426.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr17_+_73089382 | 0.13 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr7_-_100034060 | 0.13 |
ENST00000292330.2
|
PPP1R35
|
protein phosphatase 1, regulatory subunit 35 |
| chr16_+_86600857 | 0.12 |
ENST00000320354.4
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
| chr2_+_217498105 | 0.12 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr5_+_157158205 | 0.12 |
ENST00000231198.7
|
THG1L
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr17_-_76732928 | 0.12 |
ENST00000589768.1
|
CYTH1
|
cytohesin 1 |
| chr10_-_81203972 | 0.12 |
ENST00000372333.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr11_+_933555 | 0.12 |
ENST00000534485.1
|
AP2A2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chrX_+_70364667 | 0.12 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr9_+_131218408 | 0.12 |
ENST00000351030.3
ENST00000604420.1 ENST00000535026.1 ENST00000448249.3 ENST00000393527.3 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chr6_-_32821599 | 0.12 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_-_12030681 | 0.12 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr11_-_12030130 | 0.11 |
ENST00000450094.2
ENST00000534511.1 |
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr2_+_97481974 | 0.11 |
ENST00000377060.3
ENST00000305510.3 |
CNNM3
|
cyclin M3 |
| chr6_+_90272488 | 0.11 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr3_+_159481464 | 0.11 |
ENST00000467377.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr17_-_7137857 | 0.11 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2 |
| chr17_+_46132037 | 0.11 |
ENST00000582155.1
ENST00000583378.1 ENST00000536222.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr11_-_57103327 | 0.11 |
ENST00000529002.1
ENST00000278412.2 |
SSRP1
|
structure specific recognition protein 1 |
| chr7_+_114562909 | 0.11 |
ENST00000423503.1
ENST00000427207.1 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr10_-_90712520 | 0.11 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr19_-_42758040 | 0.10 |
ENST00000593944.1
|
ERF
|
Ets2 repressor factor |
| chr10_-_18948208 | 0.10 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr9_+_109625378 | 0.10 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr2_+_25016282 | 0.10 |
ENST00000260662.1
|
CENPO
|
centromere protein O |
| chr17_+_4736627 | 0.10 |
ENST00000355280.6
ENST00000347992.7 |
MINK1
|
misshapen-like kinase 1 |
| chr9_+_131218336 | 0.10 |
ENST00000372814.3
|
ODF2
|
outer dense fiber of sperm tails 2 |
| chr3_+_153839149 | 0.10 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr1_+_203595903 | 0.10 |
ENST00000367218.3
ENST00000367219.3 ENST00000391954.2 |
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr20_+_57430162 | 0.10 |
ENST00000450130.1
ENST00000349036.3 ENST00000423897.1 |
GNAS
|
GNAS complex locus |
| chr16_+_28875268 | 0.10 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr2_+_106361333 | 0.10 |
ENST00000233154.4
ENST00000451463.2 |
NCK2
|
NCK adaptor protein 2 |
| chr7_+_114562616 | 0.10 |
ENST00000448022.1
|
MDFIC
|
MyoD family inhibitor domain containing |
| chr17_-_27278445 | 0.10 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr17_-_37557846 | 0.10 |
ENST00000394294.3
ENST00000583610.1 ENST00000264658.6 |
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr10_+_99344104 | 0.09 |
ENST00000555577.1
ENST00000370649.3 |
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
| chr12_-_57030115 | 0.09 |
ENST00000379441.3
ENST00000179765.5 ENST00000551812.1 |
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr2_+_25015968 | 0.09 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chr9_+_131174024 | 0.09 |
ENST00000420034.1
ENST00000372842.1 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr6_-_29600832 | 0.08 |
ENST00000377016.4
ENST00000376977.3 ENST00000377034.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr1_+_44679159 | 0.08 |
ENST00000315913.5
ENST00000372289.2 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr3_+_10068095 | 0.08 |
ENST00000287647.3
ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2
|
Fanconi anemia, complementation group D2 |
| chr20_-_30310797 | 0.08 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr11_+_10477733 | 0.08 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr11_+_45944190 | 0.08 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr22_-_19166343 | 0.08 |
ENST00000215882.5
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr12_-_6798410 | 0.08 |
ENST00000361959.3
ENST00000436774.2 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr6_-_31620095 | 0.08 |
ENST00000424176.1
ENST00000456622.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr10_+_104404644 | 0.08 |
ENST00000462202.2
|
TRIM8
|
tripartite motif containing 8 |
| chr12_-_6798523 | 0.08 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr9_+_131219179 | 0.08 |
ENST00000372791.3
|
ODF2
|
outer dense fiber of sperm tails 2 |
| chr5_-_150460539 | 0.08 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chrX_+_48687283 | 0.08 |
ENST00000338270.1
|
ERAS
|
ES cell expressed Ras |
| chr9_+_131218698 | 0.07 |
ENST00000434106.3
ENST00000546203.1 ENST00000446274.1 ENST00000421776.2 ENST00000432065.2 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chr1_-_161014731 | 0.07 |
ENST00000368020.1
|
USF1
|
upstream transcription factor 1 |
| chr19_+_50354462 | 0.07 |
ENST00000601675.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr12_+_122516626 | 0.07 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chrX_+_51636629 | 0.07 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr20_-_30310693 | 0.07 |
ENST00000307677.4
ENST00000420653.1 |
BCL2L1
|
BCL2-like 1 |
| chr11_-_66104237 | 0.07 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr16_-_16317321 | 0.07 |
ENST00000205557.7
ENST00000575728.1 |
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
| chr9_-_34397800 | 0.07 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr1_-_205782304 | 0.07 |
ENST00000367137.3
|
SLC41A1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr20_-_30310656 | 0.07 |
ENST00000376055.4
|
BCL2L1
|
BCL2-like 1 |
| chr16_+_57844549 | 0.07 |
ENST00000564282.1
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr22_-_28197486 | 0.07 |
ENST00000302326.4
|
MN1
|
meningioma (disrupted in balanced translocation) 1 |
| chr11_-_64901978 | 0.07 |
ENST00000294256.8
ENST00000377190.3 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr15_+_40731920 | 0.07 |
ENST00000561234.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr17_+_38497640 | 0.07 |
ENST00000394086.3
|
RARA
|
retinoic acid receptor, alpha |
| chr19_+_12848299 | 0.07 |
ENST00000357332.3
|
ASNA1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr4_-_176733897 | 0.07 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr8_-_22014339 | 0.06 |
ENST00000306317.2
|
LGI3
|
leucine-rich repeat LGI family, member 3 |
| chr9_-_136242909 | 0.06 |
ENST00000371991.3
ENST00000545297.1 |
SURF4
|
surfeit 4 |
| chr1_-_156023580 | 0.06 |
ENST00000368309.3
|
UBQLN4
|
ubiquilin 4 |
| chr19_+_1383890 | 0.06 |
ENST00000539480.1
ENST00000313408.7 ENST00000414651.2 |
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) |
| chr3_+_47844399 | 0.06 |
ENST00000446256.2
ENST00000445061.1 |
DHX30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr3_+_47844615 | 0.06 |
ENST00000348968.4
|
DHX30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr1_-_156460391 | 0.06 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chr1_+_44679113 | 0.06 |
ENST00000361745.6
ENST00000446292.1 ENST00000440641.1 ENST00000436069.1 ENST00000437511.1 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr8_+_30244580 | 0.06 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr12_+_104337515 | 0.06 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr20_+_43343476 | 0.06 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr19_+_676385 | 0.06 |
ENST00000166139.4
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr9_-_98079965 | 0.06 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C |
| chr19_-_49926698 | 0.06 |
ENST00000270631.1
|
PTH2
|
parathyroid hormone 2 |
| chr17_-_17875688 | 0.06 |
ENST00000379504.3
ENST00000318094.10 ENST00000540946.1 ENST00000542206.1 ENST00000395739.4 ENST00000581396.1 ENST00000535933.1 ENST00000579586.1 |
TOM1L2
|
target of myb1-like 2 (chicken) |
| chr9_+_131219250 | 0.06 |
ENST00000470061.1
|
ODF2
|
outer dense fiber of sperm tails 2 |
| chrX_-_51812268 | 0.06 |
ENST00000486010.1
ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B
|
melanoma antigen family D, 4B |
| chr3_+_137717571 | 0.06 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr20_+_43343886 | 0.06 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr19_+_58790314 | 0.06 |
ENST00000196548.5
ENST00000608843.1 |
ZNF8
ZNF8
|
Zinc finger protein 8 zinc finger protein 8 |
| chr10_+_180643 | 0.06 |
ENST00000509513.2
ENST00000397959.3 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr6_+_111408698 | 0.06 |
ENST00000368851.5
|
SLC16A10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr17_+_26989109 | 0.06 |
ENST00000314616.6
ENST00000347486.4 |
SUPT6H
|
suppressor of Ty 6 homolog (S. cerevisiae) |
| chr6_-_31620455 | 0.06 |
ENST00000437771.1
ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6
|
BCL2-associated athanogene 6 |
| chr7_+_149411860 | 0.06 |
ENST00000486744.1
|
KRBA1
|
KRAB-A domain containing 1 |
| chr17_+_4843654 | 0.06 |
ENST00000575111.1
|
RNF167
|
ring finger protein 167 |
| chr3_+_170136642 | 0.06 |
ENST00000064724.3
ENST00000486975.1 |
CLDN11
|
claudin 11 |
| chrX_+_152240819 | 0.06 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr9_-_139372141 | 0.05 |
ENST00000313050.7
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr4_-_102268708 | 0.05 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_-_129513259 | 0.05 |
ENST00000329333.5
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr5_-_150460914 | 0.05 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr6_-_31620403 | 0.05 |
ENST00000451898.1
ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr7_-_112726393 | 0.05 |
ENST00000449591.1
ENST00000449735.1 ENST00000438062.1 ENST00000424100.1 |
GPR85
|
G protein-coupled receptor 85 |
| chr15_-_41408409 | 0.05 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chr1_+_36690011 | 0.05 |
ENST00000354618.5
ENST00000469141.2 ENST00000478853.1 |
THRAP3
|
thyroid hormone receptor associated protein 3 |
| chr17_+_7211280 | 0.05 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr19_-_6424783 | 0.05 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
| chr15_+_40861487 | 0.05 |
ENST00000315616.7
ENST00000559271.1 |
RPUSD2
|
RNA pseudouridylate synthase domain containing 2 |
| chr22_-_19165917 | 0.05 |
ENST00000451283.1
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr19_+_1495362 | 0.05 |
ENST00000395479.4
|
REEP6
|
receptor accessory protein 6 |
| chr9_+_135285579 | 0.05 |
ENST00000343036.2
ENST00000393216.2 |
C9orf171
|
chromosome 9 open reading frame 171 |
| chr6_+_90272339 | 0.05 |
ENST00000522779.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr20_+_57427765 | 0.05 |
ENST00000371100.4
|
GNAS
|
GNAS complex locus |
| chr15_-_75743915 | 0.05 |
ENST00000394949.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr11_-_75379612 | 0.05 |
ENST00000526740.1
|
MAP6
|
microtubule-associated protein 6 |
| chr19_+_42772659 | 0.05 |
ENST00000572681.2
|
CIC
|
capicua transcriptional repressor |
| chr17_+_4843679 | 0.05 |
ENST00000576229.1
|
RNF167
|
ring finger protein 167 |
| chr4_-_134070250 | 0.05 |
ENST00000505289.1
ENST00000509715.1 |
RP11-9G1.3
|
RP11-9G1.3 |
| chr11_-_64901900 | 0.05 |
ENST00000526060.1
ENST00000307289.6 ENST00000528487.1 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr22_-_30783356 | 0.05 |
ENST00000382363.3
|
RNF215
|
ring finger protein 215 |
| chr6_-_43337180 | 0.05 |
ENST00000318149.3
ENST00000361428.2 |
ZNF318
|
zinc finger protein 318 |
| chr17_-_28618867 | 0.05 |
ENST00000394819.3
ENST00000577623.1 |
BLMH
|
bleomycin hydrolase |
| chr22_-_50970566 | 0.05 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr2_-_193059634 | 0.04 |
ENST00000392314.1
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr15_+_67430339 | 0.04 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr16_+_28834303 | 0.04 |
ENST00000340394.8
ENST00000325215.6 ENST00000395547.2 ENST00000336783.4 ENST00000382686.4 ENST00000564304.1 |
ATXN2L
|
ataxin 2-like |
| chr11_+_66036004 | 0.04 |
ENST00000311481.6
ENST00000527397.1 |
RAB1B
|
RAB1B, member RAS oncogene family |
| chr11_+_76493294 | 0.04 |
ENST00000533752.1
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr12_+_57854274 | 0.04 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr11_+_45868957 | 0.04 |
ENST00000443527.2
|
CRY2
|
cryptochrome 2 (photolyase-like) |
| chr5_-_168727786 | 0.04 |
ENST00000332966.8
|
SLIT3
|
slit homolog 3 (Drosophila) |
| chr14_+_29236269 | 0.04 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr17_-_40540484 | 0.04 |
ENST00000588969.1
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr1_-_150208320 | 0.04 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_-_52486841 | 0.04 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr11_-_11643540 | 0.04 |
ENST00000227756.4
|
GALNT18
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18 |
| chr7_-_105517021 | 0.04 |
ENST00000318724.4
ENST00000419735.3 |
ATXN7L1
|
ataxin 7-like 1 |
| chr20_+_2082494 | 0.04 |
ENST00000246032.3
|
STK35
|
serine/threonine kinase 35 |
| chr19_+_36602104 | 0.04 |
ENST00000585332.1
ENST00000262637.4 |
OVOL3
|
ovo-like zinc finger 3 |
| chr10_-_97050777 | 0.04 |
ENST00000329399.6
|
PDLIM1
|
PDZ and LIM domain 1 |
| chr19_-_40919271 | 0.04 |
ENST00000291825.7
ENST00000324001.7 |
PRX
|
periaxin |
| chr19_-_39330818 | 0.04 |
ENST00000594769.1
ENST00000602021.1 |
AC104534.3
|
Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial |
| chr6_-_31620149 | 0.04 |
ENST00000435080.1
ENST00000375976.4 ENST00000441054.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr18_+_21529811 | 0.04 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr4_-_186696561 | 0.04 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_161275320 | 0.04 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chrX_-_30327495 | 0.04 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr19_+_36208877 | 0.04 |
ENST00000420124.1
ENST00000222270.7 ENST00000341701.1 |
KMT2B
|
Histone-lysine N-methyltransferase 2B |
| chr14_+_24584372 | 0.04 |
ENST00000559396.1
ENST00000558638.1 ENST00000561041.1 ENST00000559288.1 ENST00000558408.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr11_-_118789613 | 0.04 |
ENST00000532899.1
|
BCL9L
|
B-cell CLL/lymphoma 9-like |
| chr16_+_29819446 | 0.04 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr11_+_66059339 | 0.04 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chr17_-_40540377 | 0.04 |
ENST00000404395.3
ENST00000389272.3 ENST00000585517.1 ENST00000588065.1 |
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr4_-_186696425 | 0.03 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_-_86571628 | 0.03 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr3_+_137728842 | 0.03 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chrX_-_153236620 | 0.03 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr12_-_122018114 | 0.03 |
ENST00000539394.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr4_-_186696515 | 0.03 |
ENST00000456596.1
ENST00000414724.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr10_-_77161533 | 0.03 |
ENST00000535216.1
|
ZNF503
|
zinc finger protein 503 |
| chr17_+_17380294 | 0.03 |
ENST00000268711.3
ENST00000580462.1 |
MED9
|
mediator complex subunit 9 |
| chr2_-_213403565 | 0.03 |
ENST00000342788.4
ENST00000436443.1 |
ERBB4
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
| chr16_+_19183671 | 0.03 |
ENST00000562711.2
|
SYT17
|
synaptotagmin XVII |
| chr19_-_41196458 | 0.03 |
ENST00000598779.1
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr15_+_39873268 | 0.03 |
ENST00000397591.2
ENST00000260356.5 |
THBS1
|
thrombospondin 1 |
| chr11_-_65640071 | 0.03 |
ENST00000526624.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr5_+_71403061 | 0.03 |
ENST00000512974.1
ENST00000296755.7 |
MAP1B
|
microtubule-associated protein 1B |
| chr16_+_29818857 | 0.03 |
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr9_-_139371533 | 0.03 |
ENST00000290037.6
ENST00000431893.2 ENST00000371706.3 |
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr5_-_137090028 | 0.03 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr17_+_4843594 | 0.03 |
ENST00000570328.1
|
RNF167
|
ring finger protein 167 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC2_GLI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.2 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0033241 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.2 | GO:1902613 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.3 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.0 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.0 | 0.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.0 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.0 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.0 | GO:0002605 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.0 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0033270 | paranodal junction(GO:0033010) paranode region of axon(GO:0033270) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.0 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.0 | 0.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |