Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
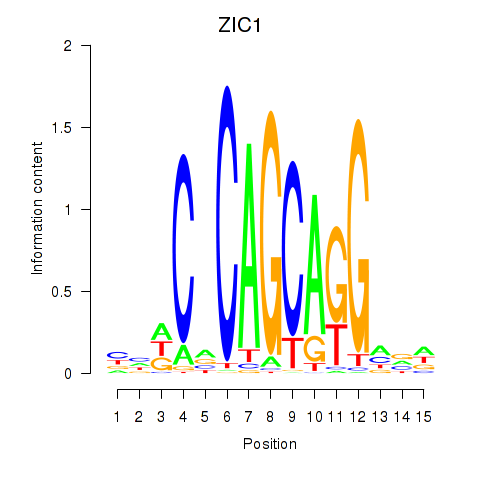
Results for ZIC1
Z-value: 0.90
Transcription factors associated with ZIC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC1
|
ENSG00000152977.5 | Zic family member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZIC1 | hg19_v2_chr3_+_147127142_147127171 | -0.73 | 1.0e-01 | Click! |
Activity profile of ZIC1 motif
Sorted Z-values of ZIC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_42931567 | 0.70 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr1_+_46668994 | 0.62 |
ENST00000371980.3
|
LURAP1
|
leucine rich adaptor protein 1 |
| chrX_+_70364667 | 0.56 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr12_-_11036844 | 0.53 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr13_-_30160925 | 0.50 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr4_+_2626988 | 0.48 |
ENST00000509050.1
|
FAM193A
|
family with sequence similarity 193, member A |
| chr17_-_3794021 | 0.48 |
ENST00000381769.2
|
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr16_-_31100284 | 0.47 |
ENST00000280606.6
|
PRSS53
|
protease, serine, 53 |
| chr12_-_11002063 | 0.46 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr11_-_57417405 | 0.46 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr13_-_45048386 | 0.45 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr16_+_771663 | 0.44 |
ENST00000568916.1
|
FAM173A
|
family with sequence similarity 173, member A |
| chr22_+_29702572 | 0.42 |
ENST00000407647.2
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr1_+_1567474 | 0.42 |
ENST00000356026.5
|
MMP23B
|
matrix metallopeptidase 23B |
| chr22_-_23922410 | 0.40 |
ENST00000249053.3
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr19_+_39647271 | 0.39 |
ENST00000599657.1
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr19_+_50169081 | 0.38 |
ENST00000246784.3
|
BCL2L12
|
BCL2-like 12 (proline rich) |
| chr11_+_208773 | 0.38 |
ENST00000528357.1
|
RIC8A
|
RIC8 guanine nucleotide exchange factor A |
| chr22_+_46972975 | 0.37 |
ENST00000431155.1
|
GRAMD4
|
GRAM domain containing 4 |
| chrX_+_53078465 | 0.37 |
ENST00000375466.2
|
GPR173
|
G protein-coupled receptor 173 |
| chr18_+_11752783 | 0.36 |
ENST00000585642.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr11_+_67777751 | 0.36 |
ENST00000316367.6
ENST00000007633.8 ENST00000342456.6 |
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr12_-_25801478 | 0.36 |
ENST00000540106.1
ENST00000445693.1 ENST00000545543.1 ENST00000542224.1 |
IFLTD1
|
intermediate filament tail domain containing 1 |
| chr17_-_67057047 | 0.35 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr1_-_1711508 | 0.34 |
ENST00000378625.1
|
NADK
|
NAD kinase |
| chr17_-_18945798 | 0.33 |
ENST00000395635.1
|
GRAP
|
GRB2-related adaptor protein |
| chr22_-_23922448 | 0.33 |
ENST00000438703.1
ENST00000330377.2 |
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr14_+_23340822 | 0.33 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr17_+_16945820 | 0.33 |
ENST00000577514.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr19_+_55987998 | 0.32 |
ENST00000591164.1
|
ZNF628
|
zinc finger protein 628 |
| chr12_-_59314246 | 0.30 |
ENST00000320743.3
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr11_+_15095108 | 0.30 |
ENST00000324229.6
ENST00000533448.1 |
CALCB
|
calcitonin-related polypeptide beta |
| chr17_+_75450099 | 0.30 |
ENST00000586433.1
|
SEPT9
|
septin 9 |
| chr1_+_17559776 | 0.30 |
ENST00000537499.1
ENST00000413717.2 ENST00000536552.1 |
PADI1
|
peptidyl arginine deiminase, type I |
| chr17_+_75450146 | 0.30 |
ENST00000585929.1
ENST00000590917.1 |
SEPT9
|
septin 9 |
| chr11_+_66824346 | 0.29 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr15_-_78112553 | 0.29 |
ENST00000562933.1
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr17_+_5981270 | 0.29 |
ENST00000571973.1
|
WSCD1
|
WSC domain containing 1 |
| chr17_+_40458130 | 0.28 |
ENST00000587646.1
|
STAT5A
|
signal transducer and activator of transcription 5A |
| chr9_-_139357413 | 0.28 |
ENST00000277537.6
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr1_-_20446020 | 0.28 |
ENST00000375105.3
|
PLA2G2D
|
phospholipase A2, group IID |
| chr18_+_11752040 | 0.27 |
ENST00000423027.3
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr11_+_66824276 | 0.27 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chrX_+_52926322 | 0.27 |
ENST00000430150.2
ENST00000452021.2 ENST00000412319.1 |
FAM156B
|
family with sequence similarity 156, member B |
| chr16_+_2255841 | 0.26 |
ENST00000301725.7
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr5_+_150406527 | 0.26 |
ENST00000520059.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr3_+_71803201 | 0.26 |
ENST00000304411.2
|
GPR27
|
G protein-coupled receptor 27 |
| chr17_+_75450075 | 0.26 |
ENST00000592951.1
|
SEPT9
|
septin 9 |
| chr16_-_745946 | 0.26 |
ENST00000562563.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr9_+_132934835 | 0.25 |
ENST00000372398.3
|
NCS1
|
neuronal calcium sensor 1 |
| chr22_+_44319619 | 0.25 |
ENST00000216180.3
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr2_+_7118755 | 0.25 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr16_+_3185125 | 0.25 |
ENST00000576416.1
|
ZNF213
|
zinc finger protein 213 |
| chr9_-_123639600 | 0.24 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr20_+_61924532 | 0.24 |
ENST00000358894.6
ENST00000326996.6 ENST00000435874.1 |
COL20A1
|
collagen, type XX, alpha 1 |
| chr2_-_97534312 | 0.24 |
ENST00000442264.1
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr19_-_3028354 | 0.24 |
ENST00000586422.1
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr19_+_34287751 | 0.24 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr7_+_29874341 | 0.24 |
ENST00000409290.1
ENST00000242140.5 |
WIPF3
|
WAS/WASL interacting protein family, member 3 |
| chr16_-_4665023 | 0.23 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr19_-_2740036 | 0.23 |
ENST00000586572.1
ENST00000269740.4 |
AC006538.4
SLC39A3
|
Uncharacterized protein solute carrier family 39 (zinc transporter), member 3 |
| chr9_+_124329336 | 0.23 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr22_-_24622080 | 0.23 |
ENST00000425408.1
|
GGT5
|
gamma-glutamyltransferase 5 |
| chr12_-_2966193 | 0.23 |
ENST00000382678.3
|
AC005841.1
|
Uncharacterized protein ENSP00000372125 |
| chr17_-_3796334 | 0.23 |
ENST00000381771.2
ENST00000348335.2 ENST00000158166.5 |
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr8_-_22526597 | 0.23 |
ENST00000519513.1
ENST00000276416.6 ENST00000520292.1 ENST00000522268.1 |
BIN3
|
bridging integrator 3 |
| chr9_-_79267432 | 0.23 |
ENST00000424866.1
|
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr3_-_53080047 | 0.23 |
ENST00000482396.1
ENST00000358080.2 ENST00000296295.6 ENST00000394752.3 |
SFMBT1
|
Scm-like with four mbt domains 1 |
| chr7_-_128045984 | 0.22 |
ENST00000470772.1
ENST00000480861.1 ENST00000496200.1 |
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chr19_+_42772659 | 0.22 |
ENST00000572681.2
|
CIC
|
capicua transcriptional repressor |
| chr10_+_118349920 | 0.21 |
ENST00000531984.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chrX_+_46772065 | 0.21 |
ENST00000455411.1
|
PHF16
|
jade family PHD finger 3 |
| chr7_-_4877677 | 0.21 |
ENST00000538469.1
|
RADIL
|
Ras association and DIL domains |
| chr6_+_90272488 | 0.21 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr21_+_18811205 | 0.21 |
ENST00000440664.1
|
C21orf37
|
chromosome 21 open reading frame 37 |
| chr1_+_44870866 | 0.21 |
ENST00000355387.2
ENST00000361799.2 |
RNF220
|
ring finger protein 220 |
| chr17_-_5372068 | 0.21 |
ENST00000572490.1
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr1_-_111991850 | 0.21 |
ENST00000411751.2
|
WDR77
|
WD repeat domain 77 |
| chr1_+_1567546 | 0.20 |
ENST00000378675.3
|
MMP23B
|
matrix metallopeptidase 23B |
| chr16_+_838614 | 0.20 |
ENST00000262315.9
ENST00000455171.2 |
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
| chr7_-_135412925 | 0.20 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr2_-_98280383 | 0.20 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr17_-_5372229 | 0.20 |
ENST00000433302.3
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr19_-_2739992 | 0.20 |
ENST00000545664.1
ENST00000589363.1 ENST00000455372.2 |
SLC39A3
|
solute carrier family 39 (zinc transporter), member 3 |
| chr19_+_10563567 | 0.20 |
ENST00000344979.3
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr15_+_41245160 | 0.20 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr5_+_140514782 | 0.19 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr3_-_50329835 | 0.19 |
ENST00000429673.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr11_+_64001962 | 0.19 |
ENST00000309422.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr16_-_4664860 | 0.19 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr16_-_2379688 | 0.19 |
ENST00000567910.1
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chr8_+_25042192 | 0.18 |
ENST00000410074.1
|
DOCK5
|
dedicator of cytokinesis 5 |
| chr15_+_77713299 | 0.18 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr11_+_64002292 | 0.18 |
ENST00000426086.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr1_+_62417957 | 0.18 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr21_+_45148735 | 0.18 |
ENST00000327574.4
|
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr19_+_42364460 | 0.17 |
ENST00000593863.1
|
RPS19
|
ribosomal protein S19 |
| chr8_+_21946681 | 0.17 |
ENST00000289921.7
|
FAM160B2
|
family with sequence similarity 160, member B2 |
| chr3_+_126243126 | 0.17 |
ENST00000319340.2
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13 |
| chr6_-_34216766 | 0.17 |
ENST00000481533.1
ENST00000468145.1 ENST00000413013.2 ENST00000394990.4 ENST00000335352.3 |
C6orf1
|
chromosome 6 open reading frame 1 |
| chr16_+_28858004 | 0.17 |
ENST00000322610.8
|
SH2B1
|
SH2B adaptor protein 1 |
| chr17_-_61778528 | 0.17 |
ENST00000584645.1
|
LIMD2
|
LIM domain containing 2 |
| chr17_-_43045439 | 0.17 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chrX_-_129299638 | 0.17 |
ENST00000535724.1
ENST00000346424.2 |
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr3_+_49027771 | 0.17 |
ENST00000475629.1
ENST00000444213.1 |
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr4_+_2627159 | 0.16 |
ENST00000382839.3
ENST00000324666.5 ENST00000545951.1 ENST00000502458.1 ENST00000505311.1 |
FAM193A
|
family with sequence similarity 193, member A |
| chr4_-_40632605 | 0.16 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr6_+_31540056 | 0.16 |
ENST00000418386.2
|
LTA
|
lymphotoxin alpha |
| chr7_-_35734176 | 0.16 |
ENST00000413517.1
ENST00000438224.1 |
HERPUD2
|
HERPUD family member 2 |
| chr1_+_24120143 | 0.16 |
ENST00000374501.1
|
LYPLA2
|
lysophospholipase II |
| chr22_+_44319648 | 0.15 |
ENST00000423180.2
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr3_+_42892458 | 0.15 |
ENST00000494619.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr9_-_123639445 | 0.15 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr6_-_168720425 | 0.14 |
ENST00000366796.3
|
DACT2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr10_-_49701686 | 0.14 |
ENST00000417247.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr16_+_30953696 | 0.14 |
ENST00000566320.2
ENST00000565939.1 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr11_-_73309228 | 0.14 |
ENST00000356467.4
ENST00000064778.4 |
FAM168A
|
family with sequence similarity 168, member A |
| chr11_+_63606373 | 0.13 |
ENST00000402010.2
ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr8_-_144623595 | 0.13 |
ENST00000262577.5
|
ZC3H3
|
zinc finger CCCH-type containing 3 |
| chrY_+_4868267 | 0.13 |
ENST00000333703.4
|
PCDH11Y
|
protocadherin 11 Y-linked |
| chrX_+_2924654 | 0.13 |
ENST00000381130.2
|
ARSH
|
arylsulfatase family, member H |
| chr7_-_73256838 | 0.13 |
ENST00000297873.4
|
WBSCR27
|
Williams Beuren syndrome chromosome region 27 |
| chr15_-_81202118 | 0.12 |
ENST00000560560.1
|
RP11-351M8.1
|
Uncharacterized protein |
| chr15_+_73976715 | 0.12 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr21_+_45209394 | 0.12 |
ENST00000497547.1
|
RRP1
|
ribosomal RNA processing 1 |
| chr3_+_50192457 | 0.12 |
ENST00000414301.1
ENST00000450338.1 |
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr3_+_14444063 | 0.12 |
ENST00000454876.2
ENST00000360861.3 ENST00000416216.2 |
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr1_-_111991908 | 0.12 |
ENST00000235090.5
|
WDR77
|
WD repeat domain 77 |
| chr19_-_6433765 | 0.12 |
ENST00000321510.6
|
SLC25A41
|
solute carrier family 25, member 41 |
| chr12_-_106477805 | 0.12 |
ENST00000553094.1
ENST00000549704.1 |
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr2_-_46769694 | 0.12 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chrX_+_18725758 | 0.12 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr17_+_60704762 | 0.12 |
ENST00000303375.5
|
MRC2
|
mannose receptor, C type 2 |
| chr1_+_65775204 | 0.11 |
ENST00000371069.4
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr1_-_89641680 | 0.11 |
ENST00000294671.2
|
GBP7
|
guanylate binding protein 7 |
| chr19_-_46142680 | 0.11 |
ENST00000245925.3
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr6_+_90272339 | 0.11 |
ENST00000522779.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr17_-_67057203 | 0.10 |
ENST00000340001.4
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr7_+_2671663 | 0.10 |
ENST00000407643.1
|
TTYH3
|
tweety family member 3 |
| chr17_-_67057114 | 0.10 |
ENST00000370732.2
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr22_-_42739533 | 0.10 |
ENST00000515426.1
|
TCF20
|
transcription factor 20 (AR1) |
| chr9_-_117420052 | 0.10 |
ENST00000423632.1
|
RP11-402G3.5
|
RP11-402G3.5 |
| chr13_-_29069232 | 0.10 |
ENST00000282397.4
ENST00000541932.1 ENST00000539099.1 |
FLT1
|
fms-related tyrosine kinase 1 |
| chr17_-_40428359 | 0.10 |
ENST00000293328.3
|
STAT5B
|
signal transducer and activator of transcription 5B |
| chr3_-_45838011 | 0.10 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr3_+_141105705 | 0.10 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr13_+_42712178 | 0.09 |
ENST00000536612.1
|
DGKH
|
diacylglycerol kinase, eta |
| chr17_+_37894570 | 0.09 |
ENST00000394211.3
|
GRB7
|
growth factor receptor-bound protein 7 |
| chr4_-_40632881 | 0.09 |
ENST00000511598.1
|
RBM47
|
RNA binding motif protein 47 |
| chr14_+_55033815 | 0.09 |
ENST00000554335.1
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr9_-_35103105 | 0.09 |
ENST00000452248.2
ENST00000356493.5 |
STOML2
|
stomatin (EPB72)-like 2 |
| chrX_-_129299847 | 0.09 |
ENST00000319908.3
ENST00000287295.3 |
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr4_-_7941596 | 0.09 |
ENST00000420658.1
ENST00000358461.2 |
AFAP1
|
actin filament associated protein 1 |
| chr19_+_38880695 | 0.08 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chrX_+_91034260 | 0.08 |
ENST00000395337.2
|
PCDH11X
|
protocadherin 11 X-linked |
| chr19_-_16582815 | 0.08 |
ENST00000455140.2
ENST00000248070.6 ENST00000594975.1 |
EPS15L1
|
epidermal growth factor receptor pathway substrate 15-like 1 |
| chr7_-_35734607 | 0.08 |
ENST00000427455.1
|
HERPUD2
|
HERPUD family member 2 |
| chr19_+_41509851 | 0.08 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr11_+_120207787 | 0.08 |
ENST00000397843.2
ENST00000356641.3 |
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr2_+_54951679 | 0.08 |
ENST00000356458.6
|
EML6
|
echinoderm microtubule associated protein like 6 |
| chr4_-_40632844 | 0.08 |
ENST00000505414.1
|
RBM47
|
RNA binding motif protein 47 |
| chr3_+_32859510 | 0.08 |
ENST00000383763.5
|
TRIM71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr3_-_45837959 | 0.08 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr1_-_113249948 | 0.08 |
ENST00000339083.7
ENST00000369642.3 |
RHOC
|
ras homolog family member C |
| chr11_+_63606477 | 0.08 |
ENST00000508192.1
ENST00000361128.5 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chrX_+_46771711 | 0.07 |
ENST00000424392.1
ENST00000397189.1 |
PHF16
|
jade family PHD finger 3 |
| chr16_-_755819 | 0.07 |
ENST00000397621.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr19_-_58609570 | 0.07 |
ENST00000600845.1
ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18
|
zinc finger and SCAN domain containing 18 |
| chr7_-_107968999 | 0.07 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chrX_+_153770421 | 0.07 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr5_-_64920115 | 0.07 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr16_-_67427389 | 0.07 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr1_+_161087873 | 0.07 |
ENST00000368009.2
ENST00000368007.4 ENST00000368008.1 ENST00000392190.5 |
NIT1
|
nitrilase 1 |
| chrX_+_118425471 | 0.07 |
ENST00000428222.1
|
RP5-1139I1.1
|
RP5-1139I1.1 |
| chr2_+_232457569 | 0.07 |
ENST00000313965.2
|
C2orf57
|
chromosome 2 open reading frame 57 |
| chr11_+_7598239 | 0.06 |
ENST00000525597.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr10_-_33623564 | 0.06 |
ENST00000374875.1
ENST00000374822.4 |
NRP1
|
neuropilin 1 |
| chr4_-_40632757 | 0.06 |
ENST00000511902.1
ENST00000505220.1 |
RBM47
|
RNA binding motif protein 47 |
| chr1_-_113249678 | 0.06 |
ENST00000369633.2
ENST00000425265.2 ENST00000369632.2 ENST00000436685.2 |
RHOC
|
ras homolog family member C |
| chr2_-_99552620 | 0.06 |
ENST00000428096.1
ENST00000397899.2 ENST00000420294.1 |
KIAA1211L
|
KIAA1211-like |
| chr1_-_161087802 | 0.06 |
ENST00000368010.3
|
PFDN2
|
prefoldin subunit 2 |
| chr7_+_39017504 | 0.06 |
ENST00000403058.1
|
POU6F2
|
POU class 6 homeobox 2 |
| chr16_-_11485922 | 0.06 |
ENST00000599216.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr1_-_43833628 | 0.06 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr22_+_21369316 | 0.06 |
ENST00000413302.2
ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr19_-_50168962 | 0.06 |
ENST00000599223.1
ENST00000593922.1 ENST00000600022.1 ENST00000596765.1 ENST00000599144.1 ENST00000596822.1 ENST00000598108.1 ENST00000601373.1 ENST00000595034.1 ENST00000601291.1 |
IRF3
|
interferon regulatory factor 3 |
| chr18_-_52989217 | 0.06 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr15_+_73976545 | 0.06 |
ENST00000318443.5
ENST00000537340.2 ENST00000318424.5 |
CD276
|
CD276 molecule |
| chr17_-_53809473 | 0.06 |
ENST00000575734.1
|
TMEM100
|
transmembrane protein 100 |
| chr19_-_50169064 | 0.06 |
ENST00000593337.1
ENST00000598808.1 ENST00000600453.1 ENST00000593818.1 ENST00000597198.1 ENST00000601809.1 ENST00000377139.3 |
IRF3
|
interferon regulatory factor 3 |
| chr15_-_43863695 | 0.06 |
ENST00000439195.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr22_-_30953587 | 0.05 |
ENST00000453479.1
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr19_+_35940486 | 0.05 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr6_+_28109703 | 0.04 |
ENST00000457389.2
ENST00000330236.6 |
ZKSCAN8
|
zinc finger with KRAB and SCAN domains 8 |
| chr1_-_113249734 | 0.04 |
ENST00000484054.3
ENST00000369636.2 ENST00000369637.1 ENST00000285735.2 ENST00000369638.2 |
RHOC
|
ras homolog family member C |
| chr4_-_40632140 | 0.04 |
ENST00000514782.1
|
RBM47
|
RNA binding motif protein 47 |
| chr17_-_6947225 | 0.04 |
ENST00000574600.1
ENST00000308009.1 ENST00000447225.1 |
SLC16A11
|
solute carrier family 16, member 11 |
| chrX_+_68835911 | 0.04 |
ENST00000525810.1
ENST00000527388.1 ENST00000374553.2 ENST00000374552.4 ENST00000338901.3 ENST00000524573.1 |
EDA
|
ectodysplasin A |
| chr17_-_5372271 | 0.04 |
ENST00000225296.3
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr4_-_175750364 | 0.04 |
ENST00000340217.5
ENST00000274093.3 |
GLRA3
|
glycine receptor, alpha 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.6 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 0.3 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.2 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.1 | 0.4 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.3 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 | 0.4 | GO:0000255 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) |
| 0.1 | 0.5 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.3 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.2 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.1 | 0.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.3 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.2 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.2 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.2 | GO:0060266 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.4 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.3 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.4 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.9 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.0 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.4 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.4 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.4 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0051493 | regulation of cytoskeleton organization(GO:0051493) |
| 0.0 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.0 | 0.2 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.7 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.0 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.4 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.2 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.1 | 0.5 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.4 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |