Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
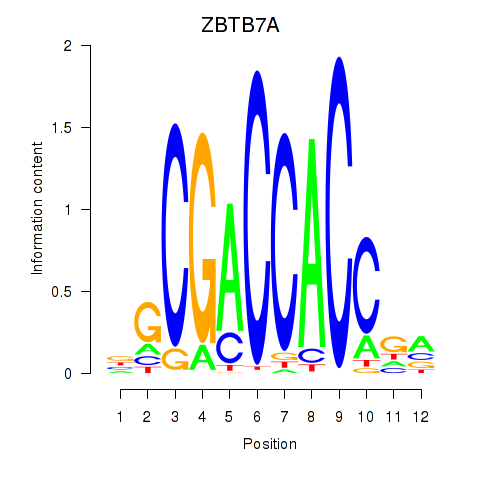
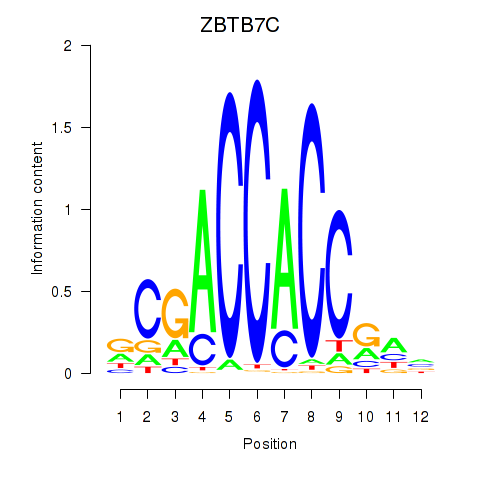
Results for ZBTB7A_ZBTB7C
Z-value: 0.64
Transcription factors associated with ZBTB7A_ZBTB7C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB7A
|
ENSG00000178951.4 | zinc finger and BTB domain containing 7A |
|
ZBTB7C
|
ENSG00000184828.5 | zinc finger and BTB domain containing 7C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB7A | hg19_v2_chr19_-_4065730_4065909 | -0.70 | 1.2e-01 | Click! |
| ZBTB7C | hg19_v2_chr18_-_45935663_45935793 | 0.08 | 8.8e-01 | Click! |
Activity profile of ZBTB7A_ZBTB7C motif
Sorted Z-values of ZBTB7A_ZBTB7C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_42631529 | 0.57 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr21_+_38338737 | 0.46 |
ENST00000430068.1
|
AP000704.5
|
AP000704.5 |
| chr15_+_27112948 | 0.44 |
ENST00000555060.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr2_-_235405168 | 0.39 |
ENST00000339728.3
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr18_+_74207477 | 0.34 |
ENST00000532511.1
|
RP11-17M16.1
|
uncharacterized protein LOC400658 |
| chr7_-_94285472 | 0.34 |
ENST00000437425.2
ENST00000447873.1 ENST00000415788.2 |
SGCE
|
sarcoglycan, epsilon |
| chr13_+_20532900 | 0.30 |
ENST00000382871.2
|
ZMYM2
|
zinc finger, MYM-type 2 |
| chr7_-_94285402 | 0.27 |
ENST00000428696.2
ENST00000445866.2 |
SGCE
|
sarcoglycan, epsilon |
| chr9_+_96717821 | 0.24 |
ENST00000454594.1
|
RP11-231K24.2
|
RP11-231K24.2 |
| chr1_-_111506562 | 0.24 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr3_-_71803474 | 0.23 |
ENST00000448225.1
ENST00000496214.2 |
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr7_+_77167343 | 0.23 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr7_+_77167376 | 0.22 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr3_+_108308559 | 0.22 |
ENST00000486815.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr5_-_114515734 | 0.22 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr9_-_123476612 | 0.21 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr8_+_17354617 | 0.21 |
ENST00000470360.1
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr16_+_53164833 | 0.21 |
ENST00000564845.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr5_-_145214893 | 0.20 |
ENST00000394450.2
|
PRELID2
|
PRELI domain containing 2 |
| chr10_+_60145155 | 0.20 |
ENST00000373895.3
|
TFAM
|
transcription factor A, mitochondrial |
| chr8_+_17354587 | 0.20 |
ENST00000494857.1
ENST00000522656.1 |
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr5_+_102455968 | 0.20 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr6_+_64282447 | 0.19 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr10_+_60144782 | 0.19 |
ENST00000487519.1
|
TFAM
|
transcription factor A, mitochondrial |
| chr14_-_54955721 | 0.18 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr20_+_36149602 | 0.18 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr9_+_74526384 | 0.18 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr13_-_30881134 | 0.18 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr13_-_30880979 | 0.18 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr3_-_71803917 | 0.17 |
ENST00000421769.2
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr7_-_94285511 | 0.17 |
ENST00000265735.7
|
SGCE
|
sarcoglycan, epsilon |
| chr2_+_46769798 | 0.17 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr5_+_72861560 | 0.17 |
ENST00000296792.4
ENST00000509005.1 ENST00000543251.1 ENST00000508686.1 ENST00000508491.1 |
UTP15
|
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) |
| chr14_-_22005197 | 0.17 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr8_+_27629459 | 0.16 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr9_-_127905736 | 0.16 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr3_+_179040767 | 0.16 |
ENST00000496856.1
ENST00000491818.1 |
ZNF639
|
zinc finger protein 639 |
| chr14_+_64970427 | 0.15 |
ENST00000553583.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr13_+_96204961 | 0.15 |
ENST00000299339.2
|
CLDN10
|
claudin 10 |
| chr19_+_10197463 | 0.15 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chrX_+_133507327 | 0.15 |
ENST00000332070.3
ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6
|
PHD finger protein 6 |
| chr3_-_136471204 | 0.15 |
ENST00000480733.1
ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1
|
stromal antigen 1 |
| chr4_-_56412713 | 0.15 |
ENST00000435527.2
|
CLOCK
|
clock circadian regulator |
| chrX_+_133507283 | 0.15 |
ENST00000370803.3
|
PHF6
|
PHD finger protein 6 |
| chr14_+_59951161 | 0.14 |
ENST00000261247.9
ENST00000425728.2 ENST00000556985.1 ENST00000554271.1 ENST00000554795.1 |
JKAMP
|
JNK1/MAPK8-associated membrane protein |
| chr19_-_48673465 | 0.14 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr17_+_29158962 | 0.14 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chr13_+_20532848 | 0.14 |
ENST00000382874.2
|
ZMYM2
|
zinc finger, MYM-type 2 |
| chr14_-_22005062 | 0.14 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chrX_-_119603138 | 0.14 |
ENST00000200639.4
ENST00000371335.4 ENST00000538785.1 ENST00000434600.2 |
LAMP2
|
lysosomal-associated membrane protein 2 |
| chr19_-_39832563 | 0.13 |
ENST00000599274.1
|
CTC-246B18.10
|
CTC-246B18.10 |
| chr10_+_14880364 | 0.13 |
ENST00000441647.1
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr2_+_27505260 | 0.13 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr22_+_38035623 | 0.13 |
ENST00000336738.5
ENST00000442465.2 |
SH3BP1
|
SH3-domain binding protein 1 |
| chr2_+_149402009 | 0.13 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr8_-_101964231 | 0.13 |
ENST00000521309.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr6_+_87865262 | 0.13 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr16_+_2014993 | 0.12 |
ENST00000564014.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr14_-_54955376 | 0.12 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr8_-_101963677 | 0.12 |
ENST00000395956.3
ENST00000395953.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr22_+_38035459 | 0.11 |
ENST00000357436.4
|
SH3BP1
|
SH3-domain binding protein 1 |
| chr8_-_101964265 | 0.11 |
ENST00000395958.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr6_-_109703634 | 0.11 |
ENST00000324953.5
ENST00000310786.4 ENST00000275080.7 ENST00000413644.2 |
CD164
|
CD164 molecule, sialomucin |
| chr6_+_73331520 | 0.11 |
ENST00000342056.2
ENST00000355194.4 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr10_+_49514698 | 0.11 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr4_+_95373037 | 0.11 |
ENST00000359265.4
ENST00000437932.1 ENST00000380180.3 ENST00000318007.5 ENST00000450793.1 ENST00000538141.1 ENST00000317968.4 ENST00000512274.1 ENST00000503974.1 ENST00000504489.1 ENST00000542407.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr20_-_58508359 | 0.10 |
ENST00000446834.1
|
SYCP2
|
synaptonemal complex protein 2 |
| chr17_-_63556414 | 0.10 |
ENST00000585045.1
|
AXIN2
|
axin 2 |
| chr14_+_64970662 | 0.10 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr10_-_14880002 | 0.10 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr6_-_122792919 | 0.10 |
ENST00000339697.4
|
SERINC1
|
serine incorporator 1 |
| chr1_+_167905894 | 0.10 |
ENST00000367843.3
ENST00000432587.2 ENST00000312263.6 |
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr12_-_120805872 | 0.10 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr7_+_26191809 | 0.09 |
ENST00000056233.3
|
NFE2L3
|
nuclear factor, erythroid 2-like 3 |
| chr20_-_58508702 | 0.09 |
ENST00000357552.3
ENST00000425931.1 |
SYCP2
|
synaptonemal complex protein 2 |
| chr5_+_76506706 | 0.09 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr2_-_153032484 | 0.09 |
ENST00000263904.4
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr8_-_101963482 | 0.09 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr16_+_610407 | 0.09 |
ENST00000409413.3
|
C16orf11
|
chromosome 16 open reading frame 11 |
| chrX_-_122866874 | 0.09 |
ENST00000245838.8
ENST00000355725.4 |
THOC2
|
THO complex 2 |
| chr3_+_179322481 | 0.09 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr4_-_76598326 | 0.09 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr4_+_15005391 | 0.09 |
ENST00000507071.1
ENST00000345451.3 ENST00000259997.5 ENST00000382395.3 ENST00000382401.3 |
CPEB2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr19_-_5567842 | 0.08 |
ENST00000587632.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr2_+_191513587 | 0.08 |
ENST00000416973.1
ENST00000426601.1 |
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr20_-_23402028 | 0.08 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr3_-_108308241 | 0.08 |
ENST00000295746.8
|
KIAA1524
|
KIAA1524 |
| chr12_-_56224546 | 0.08 |
ENST00000357606.3
ENST00000547445.1 |
DNAJC14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr9_+_74764340 | 0.08 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr8_+_22298578 | 0.08 |
ENST00000240139.5
ENST00000289963.8 ENST00000397775.3 |
PPP3CC
|
protein phosphatase 3, catalytic subunit, gamma isozyme |
| chr10_+_70661014 | 0.08 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr2_+_191513959 | 0.08 |
ENST00000337386.5
ENST00000357215.5 |
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr14_+_70346125 | 0.08 |
ENST00000361956.3
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr2_-_64246206 | 0.08 |
ENST00000409558.4
ENST00000272322.4 |
VPS54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr3_-_15469045 | 0.08 |
ENST00000450816.2
|
METTL6
|
methyltransferase like 6 |
| chr3_-_185542817 | 0.08 |
ENST00000382199.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr2_+_48667983 | 0.08 |
ENST00000449090.2
|
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr2_+_191513789 | 0.08 |
ENST00000409581.1
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr9_-_77643307 | 0.08 |
ENST00000376834.3
ENST00000376830.3 |
C9orf41
|
chromosome 9 open reading frame 41 |
| chr4_-_83719983 | 0.08 |
ENST00000319540.4
|
SCD5
|
stearoyl-CoA desaturase 5 |
| chr2_+_136289030 | 0.08 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr7_-_26904317 | 0.07 |
ENST00000345317.2
|
SKAP2
|
src kinase associated phosphoprotein 2 |
| chr3_+_108308513 | 0.07 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr12_-_56221330 | 0.07 |
ENST00000546837.1
|
RP11-762I7.5
|
Uncharacterized protein |
| chr9_-_74525658 | 0.07 |
ENST00000333421.6
|
ABHD17B
|
abhydrolase domain containing 17B |
| chr4_-_76598296 | 0.07 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr3_+_111578640 | 0.07 |
ENST00000393925.3
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr2_+_47630108 | 0.07 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr8_-_30890710 | 0.07 |
ENST00000523392.1
|
PURG
|
purine-rich element binding protein G |
| chr15_-_59949693 | 0.07 |
ENST00000396063.1
ENST00000396064.3 ENST00000484743.1 ENST00000559706.1 ENST00000396060.2 |
GTF2A2
|
general transcription factor IIA, 2, 12kDa |
| chrX_+_133507389 | 0.07 |
ENST00000370800.4
|
PHF6
|
PHD finger protein 6 |
| chr6_-_166755995 | 0.07 |
ENST00000361731.3
|
SFT2D1
|
SFT2 domain containing 1 |
| chr11_-_117747607 | 0.07 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr3_-_100120223 | 0.07 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr19_-_49222956 | 0.07 |
ENST00000599703.1
ENST00000318083.6 ENST00000419611.1 ENST00000377367.3 |
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr10_+_70320413 | 0.07 |
ENST00000373644.4
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr14_-_22005018 | 0.07 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr1_-_167906277 | 0.07 |
ENST00000271373.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr2_+_47630255 | 0.07 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chr21_-_45079341 | 0.07 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr3_-_179322436 | 0.07 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr3_+_23986748 | 0.06 |
ENST00000312521.4
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr8_+_22298767 | 0.06 |
ENST00000522000.1
|
PPP3CC
|
protein phosphatase 3, catalytic subunit, gamma isozyme |
| chr5_+_78532003 | 0.06 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr8_+_26240414 | 0.06 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr3_+_179322573 | 0.06 |
ENST00000493866.1
ENST00000472629.1 ENST00000482604.1 |
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr1_+_162467595 | 0.06 |
ENST00000538489.1
ENST00000489294.1 |
UHMK1
|
U2AF homology motif (UHM) kinase 1 |
| chr1_+_169075554 | 0.06 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr12_+_13197218 | 0.06 |
ENST00000197268.8
|
KIAA1467
|
KIAA1467 |
| chr5_-_2751762 | 0.06 |
ENST00000302057.5
ENST00000382611.6 |
IRX2
|
iroquois homeobox 2 |
| chr8_-_74884482 | 0.06 |
ENST00000520242.1
ENST00000519082.1 |
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr15_+_36871983 | 0.06 |
ENST00000437989.2
ENST00000569302.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr12_+_107349497 | 0.06 |
ENST00000548125.1
ENST00000280756.4 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr2_-_64371546 | 0.06 |
ENST00000358912.4
|
PELI1
|
pellino E3 ubiquitin protein ligase 1 |
| chrX_-_151143140 | 0.06 |
ENST00000393914.3
ENST00000370328.3 ENST00000370325.1 |
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr15_-_83876758 | 0.06 |
ENST00000299633.4
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr3_+_111578027 | 0.06 |
ENST00000431670.2
ENST00000412622.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr2_+_48667898 | 0.06 |
ENST00000281394.4
ENST00000294952.8 |
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr7_+_116312411 | 0.06 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr3_-_72496035 | 0.06 |
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein |
| chr12_+_22199108 | 0.06 |
ENST00000229329.2
|
CMAS
|
cytidine monophosphate N-acetylneuraminic acid synthetase |
| chr5_-_33984741 | 0.06 |
ENST00000382102.3
ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2
|
solute carrier family 45, member 2 |
| chrX_-_109561294 | 0.06 |
ENST00000372059.2
ENST00000262844.5 |
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr3_-_33759541 | 0.06 |
ENST00000468888.2
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr17_-_3704500 | 0.06 |
ENST00000263087.4
|
ITGAE
|
integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) |
| chr4_-_83719884 | 0.06 |
ENST00000282709.4
ENST00000273908.4 |
SCD5
|
stearoyl-CoA desaturase 5 |
| chr12_+_132312931 | 0.06 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr4_-_83351005 | 0.05 |
ENST00000295470.5
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr19_-_30199516 | 0.05 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr2_+_58655520 | 0.05 |
ENST00000455219.3
ENST00000449448.2 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr9_-_74525847 | 0.05 |
ENST00000377041.2
|
ABHD17B
|
abhydrolase domain containing 17B |
| chr5_-_137878887 | 0.05 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr2_+_242255275 | 0.05 |
ENST00000391971.2
|
SEPT2
|
septin 2 |
| chr14_+_59655369 | 0.05 |
ENST00000360909.3
ENST00000351081.1 ENST00000556135.1 |
DAAM1
|
dishevelled associated activator of morphogenesis 1 |
| chr10_+_14880157 | 0.05 |
ENST00000378372.3
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr3_-_185542761 | 0.05 |
ENST00000457616.2
ENST00000346192.3 |
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr8_-_74884459 | 0.05 |
ENST00000522337.1
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr3_-_149470229 | 0.05 |
ENST00000473414.1
|
COMMD2
|
COMM domain containing 2 |
| chr8_-_74884341 | 0.05 |
ENST00000284811.8
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr17_+_61600682 | 0.05 |
ENST00000581784.1
ENST00000456941.2 ENST00000580652.1 ENST00000314672.5 ENST00000583023.1 |
KCNH6
|
potassium voltage-gated channel, subfamily H (eag-related), member 6 |
| chr16_+_66400533 | 0.05 |
ENST00000341529.3
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr3_-_33759699 | 0.05 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr6_-_109703600 | 0.05 |
ENST00000512821.1
|
CD164
|
CD164 molecule, sialomucin |
| chrY_+_15815447 | 0.05 |
ENST00000284856.3
|
TMSB4Y
|
thymosin beta 4, Y-linked |
| chr17_-_8534031 | 0.05 |
ENST00000411957.1
ENST00000396239.1 ENST00000379980.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr1_+_205538105 | 0.05 |
ENST00000367147.4
ENST00000539267.1 |
MFSD4
|
major facilitator superfamily domain containing 4 |
| chr8_-_38853990 | 0.05 |
ENST00000456845.2
ENST00000397070.2 ENST00000517872.1 ENST00000412303.1 ENST00000456397.2 |
TM2D2
|
TM2 domain containing 2 |
| chrX_+_152783131 | 0.05 |
ENST00000349466.2
ENST00000370186.1 |
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr4_-_16085340 | 0.05 |
ENST00000508167.1
|
PROM1
|
prominin 1 |
| chr14_-_24584138 | 0.05 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr1_-_43205811 | 0.04 |
ENST00000372539.3
ENST00000296387.1 ENST00000539749.1 |
CLDN19
|
claudin 19 |
| chr1_+_167906056 | 0.04 |
ENST00000367840.3
|
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr12_-_107487604 | 0.04 |
ENST00000008527.5
|
CRY1
|
cryptochrome 1 (photolyase-like) |
| chr1_+_89246647 | 0.04 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr14_-_64970494 | 0.04 |
ENST00000608382.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr17_-_8868991 | 0.04 |
ENST00000447110.1
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr12_-_96184533 | 0.04 |
ENST00000343702.4
ENST00000344911.4 |
NTN4
|
netrin 4 |
| chr4_+_39184024 | 0.04 |
ENST00000399820.3
ENST00000509560.1 ENST00000512112.1 ENST00000288634.7 ENST00000506503.1 |
WDR19
|
WD repeat domain 19 |
| chr2_-_136288740 | 0.04 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr2_+_37311588 | 0.04 |
ENST00000409774.1
ENST00000608836.1 |
GPATCH11
|
G patch domain containing 11 |
| chr6_+_125474992 | 0.04 |
ENST00000528193.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr7_-_151107106 | 0.04 |
ENST00000334493.6
|
WDR86
|
WD repeat domain 86 |
| chr5_-_72861484 | 0.04 |
ENST00000296785.3
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr19_-_56988677 | 0.04 |
ENST00000504904.3
ENST00000292069.6 |
ZNF667
|
zinc finger protein 667 |
| chr4_-_47916543 | 0.04 |
ENST00000507489.1
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1 |
| chr12_+_50355647 | 0.04 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr19_-_663147 | 0.04 |
ENST00000606702.1
|
RNF126
|
ring finger protein 126 |
| chr17_+_6926339 | 0.04 |
ENST00000293805.5
|
BCL6B
|
B-cell CLL/lymphoma 6, member B |
| chr3_-_179322416 | 0.04 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr6_-_41715128 | 0.04 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr8_-_74884511 | 0.03 |
ENST00000518127.1
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr2_+_37311645 | 0.03 |
ENST00000281932.5
|
GPATCH11
|
G patch domain containing 11 |
| chr5_+_138210919 | 0.03 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr2_+_149402553 | 0.03 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr14_+_70233810 | 0.03 |
ENST00000394366.2
ENST00000553548.1 ENST00000553369.1 ENST00000557154.1 ENST00000451983.2 ENST00000553635.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chr16_-_49315731 | 0.03 |
ENST00000219197.6
|
CBLN1
|
cerebellin 1 precursor |
| chr14_-_51411194 | 0.03 |
ENST00000544180.2
|
PYGL
|
phosphorylase, glycogen, liver |
| chr10_+_70091812 | 0.03 |
ENST00000265866.7
|
HNRNPH3
|
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr15_-_62352659 | 0.03 |
ENST00000249837.3
|
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr17_-_7080883 | 0.03 |
ENST00000570576.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB7A_ZBTB7C
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061461 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.1 | 0.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.3 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.2 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.1 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0006311 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.0 | 0.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.5 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0032759 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.0 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.1 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.0 | 0.1 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.3 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.4 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005289 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.1 | 0.4 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.1 | GO:0032181 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.0 | 0.2 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.0 | GO:0008184 | nucleobase binding(GO:0002054) purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |