Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
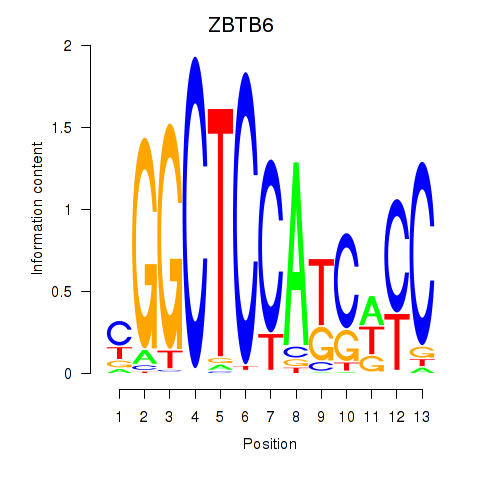
Results for ZBTB6
Z-value: 1.57
Transcription factors associated with ZBTB6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB6
|
ENSG00000186130.4 | zinc finger and BTB domain containing 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB6 | hg19_v2_chr9_-_125675576_125675612 | -0.68 | 1.4e-01 | Click! |
Activity profile of ZBTB6 motif
Sorted Z-values of ZBTB6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_1429010 | 0.97 |
ENST00000513783.1
|
UNKL
|
unkempt family zinc finger-like |
| chr18_+_20715416 | 0.87 |
ENST00000580153.1
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr12_-_58145889 | 0.81 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr16_-_28503327 | 0.64 |
ENST00000535392.1
ENST00000395653.4 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr6_+_157099036 | 0.64 |
ENST00000350026.5
ENST00000346085.5 ENST00000367148.1 ENST00000275248.4 |
ARID1B
|
AT rich interactive domain 1B (SWI1-like) |
| chr16_-_28503080 | 0.62 |
ENST00000565316.1
ENST00000565778.1 ENST00000357857.9 ENST00000568558.1 ENST00000357806.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr17_+_40688190 | 0.61 |
ENST00000225927.2
|
NAGLU
|
N-acetylglucosaminidase, alpha |
| chr12_+_48147699 | 0.60 |
ENST00000548498.1
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr11_-_66313699 | 0.59 |
ENST00000526986.1
ENST00000310442.3 |
ZDHHC24
|
zinc finger, DHHC-type containing 24 |
| chr10_+_81892477 | 0.58 |
ENST00000372263.3
|
PLAC9
|
placenta-specific 9 |
| chr22_-_23922410 | 0.58 |
ENST00000249053.3
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr11_+_826136 | 0.56 |
ENST00000528315.1
ENST00000533803.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr3_+_112929850 | 0.54 |
ENST00000464546.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr19_-_4717835 | 0.54 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr2_-_135476552 | 0.54 |
ENST00000281924.6
|
TMEM163
|
transmembrane protein 163 |
| chr5_-_151066514 | 0.52 |
ENST00000538026.1
ENST00000522348.1 ENST00000521569.1 |
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr6_-_33168391 | 0.52 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr19_+_4304685 | 0.51 |
ENST00000601006.1
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr16_-_28503357 | 0.50 |
ENST00000333496.9
ENST00000561505.1 ENST00000567963.1 ENST00000354630.5 ENST00000355477.5 ENST00000357076.5 ENST00000565688.1 ENST00000359984.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr16_-_4039001 | 0.50 |
ENST00000576936.1
|
ADCY9
|
adenylate cyclase 9 |
| chr17_+_77893135 | 0.50 |
ENST00000574526.1
ENST00000572353.1 |
RP11-353N14.4
|
RP11-353N14.4 |
| chr20_-_34287220 | 0.50 |
ENST00000306750.3
|
NFS1
|
NFS1 cysteine desulfurase |
| chr16_-_4664382 | 0.50 |
ENST00000591113.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr8_+_63161491 | 0.49 |
ENST00000523211.1
ENST00000524201.1 |
NKAIN3
|
Na+/K+ transporting ATPase interacting 3 |
| chr12_-_120189900 | 0.49 |
ENST00000546026.1
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr15_+_85144217 | 0.49 |
ENST00000540936.1
ENST00000448803.2 ENST00000546275.1 ENST00000546148.1 ENST00000442073.3 ENST00000334141.3 ENST00000358472.3 ENST00000502939.2 ENST00000379358.3 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr2_+_27282134 | 0.48 |
ENST00000441931.1
|
AGBL5
|
ATP/GTP binding protein-like 5 |
| chr1_+_26856236 | 0.48 |
ENST00000374168.2
ENST00000374166.4 |
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr17_-_7080883 | 0.47 |
ENST00000570576.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr16_+_66461175 | 0.47 |
ENST00000536005.2
ENST00000299694.8 ENST00000561796.1 |
BEAN1
|
brain expressed, associated with NEDD4, 1 |
| chr1_+_43996518 | 0.46 |
ENST00000359947.4
ENST00000438120.1 |
PTPRF
|
protein tyrosine phosphatase, receptor type, F |
| chr14_+_100594914 | 0.46 |
ENST00000554695.1
|
EVL
|
Enah/Vasp-like |
| chr15_-_41805994 | 0.45 |
ENST00000561619.1
ENST00000263800.6 ENST00000355166.5 ENST00000453182.2 |
LTK
|
leukocyte receptor tyrosine kinase |
| chr19_+_59055814 | 0.45 |
ENST00000594806.1
ENST00000253024.5 ENST00000341753.6 |
TRIM28
|
tripartite motif containing 28 |
| chr8_+_99956662 | 0.45 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr1_-_26633480 | 0.45 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chr14_-_61191049 | 0.44 |
ENST00000556952.3
|
SIX4
|
SIX homeobox 4 |
| chr19_+_51815102 | 0.44 |
ENST00000270642.8
|
IGLON5
|
IgLON family member 5 |
| chr22_-_23922448 | 0.43 |
ENST00000438703.1
ENST00000330377.2 |
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr16_+_1756162 | 0.43 |
ENST00000250894.4
ENST00000356010.5 |
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr1_-_26633067 | 0.43 |
ENST00000421827.2
ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11
|
UBX domain protein 11 |
| chr2_-_68547061 | 0.43 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr16_-_86588627 | 0.42 |
ENST00000565482.1
ENST00000564364.1 ENST00000561989.1 ENST00000543303.2 ENST00000381214.5 ENST00000360900.6 ENST00000322911.6 ENST00000546093.1 ENST00000569000.1 ENST00000562994.1 ENST00000561522.1 |
MTHFSD
|
methenyltetrahydrofolate synthetase domain containing |
| chr3_+_150126101 | 0.42 |
ENST00000361875.3
ENST00000361136.2 |
TSC22D2
|
TSC22 domain family, member 2 |
| chr22_+_31489344 | 0.42 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr22_+_38004942 | 0.42 |
ENST00000439161.1
ENST00000449944.1 ENST00000411501.1 ENST00000453208.1 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr15_+_41186609 | 0.42 |
ENST00000220509.5
|
VPS18
|
vacuolar protein sorting 18 homolog (S. cerevisiae) |
| chr8_-_72756667 | 0.41 |
ENST00000325509.4
|
MSC
|
musculin |
| chr1_-_204165610 | 0.41 |
ENST00000367194.4
|
KISS1
|
KiSS-1 metastasis-suppressor |
| chr1_+_53793885 | 0.40 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr22_-_31688431 | 0.40 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr14_-_69262947 | 0.40 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr17_-_48278983 | 0.40 |
ENST00000225964.5
|
COL1A1
|
collagen, type I, alpha 1 |
| chr16_-_787771 | 0.40 |
ENST00000568545.1
|
NARFL
|
nuclear prelamin A recognition factor-like |
| chr16_-_23724518 | 0.40 |
ENST00000457008.2
|
ERN2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr20_-_30458432 | 0.39 |
ENST00000375966.4
ENST00000278979.3 |
DUSP15
|
dual specificity phosphatase 15 |
| chr22_+_38004473 | 0.39 |
ENST00000414350.3
ENST00000343632.4 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr15_+_101256294 | 0.39 |
ENST00000559755.1
|
RP11-66B24.5
|
RP11-66B24.5 |
| chr12_+_113659234 | 0.39 |
ENST00000551096.1
ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1
|
two pore segment channel 1 |
| chr7_-_150864635 | 0.38 |
ENST00000297537.4
|
GBX1
|
gastrulation brain homeobox 1 |
| chr19_-_39926268 | 0.38 |
ENST00000599705.1
|
RPS16
|
ribosomal protein S16 |
| chr6_+_36165133 | 0.38 |
ENST00000446974.1
ENST00000454960.1 |
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr2_+_74229812 | 0.38 |
ENST00000305799.7
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chr11_+_65383227 | 0.38 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr1_+_44444865 | 0.38 |
ENST00000372324.1
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr19_-_45663408 | 0.38 |
ENST00000317951.4
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1 |
| chrX_-_106960285 | 0.38 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr17_-_17140436 | 0.37 |
ENST00000285071.4
ENST00000389169.5 ENST00000417064.1 |
FLCN
|
folliculin |
| chr17_+_7155556 | 0.37 |
ENST00000570500.1
ENST00000574993.1 ENST00000396628.2 ENST00000573657.1 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr2_+_166095898 | 0.37 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr1_-_31902614 | 0.37 |
ENST00000596131.1
|
AC114494.1
|
HCG1787699; Uncharacterized protein |
| chr16_-_67450325 | 0.37 |
ENST00000348579.2
|
ZDHHC1
|
zinc finger, DHHC-type containing 1 |
| chr10_+_101292684 | 0.36 |
ENST00000344586.7
|
NKX2-3
|
NK2 homeobox 3 |
| chr22_-_31688381 | 0.36 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr19_+_41698927 | 0.36 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr2_-_27531313 | 0.36 |
ENST00000296099.2
|
UCN
|
urocortin |
| chr7_-_642261 | 0.36 |
ENST00000400758.2
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr14_-_73493825 | 0.36 |
ENST00000318876.5
ENST00000556143.1 |
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr16_-_31021717 | 0.35 |
ENST00000565419.1
|
STX1B
|
syntaxin 1B |
| chr16_+_86600857 | 0.35 |
ENST00000320354.4
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
| chr7_+_97736197 | 0.35 |
ENST00000297293.5
|
LMTK2
|
lemur tyrosine kinase 2 |
| chr22_+_38004832 | 0.35 |
ENST00000405147.3
ENST00000429218.1 ENST00000325180.8 ENST00000337437.4 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr19_+_2164126 | 0.35 |
ENST00000398665.3
|
DOT1L
|
DOT1-like histone H3K79 methyltransferase |
| chr10_-_15413035 | 0.35 |
ENST00000378116.4
ENST00000455654.1 |
FAM171A1
|
family with sequence similarity 171, member A1 |
| chr15_+_41186637 | 0.34 |
ENST00000558474.1
|
VPS18
|
vacuolar protein sorting 18 homolog (S. cerevisiae) |
| chr5_+_177557997 | 0.34 |
ENST00000313386.4
ENST00000515098.1 ENST00000542098.1 ENST00000502814.1 ENST00000507457.1 ENST00000508647.1 |
RMND5B
|
required for meiotic nuclear division 5 homolog B (S. cerevisiae) |
| chr6_+_63921399 | 0.34 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr16_+_57679859 | 0.34 |
ENST00000569494.1
ENST00000566169.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr17_-_7518145 | 0.34 |
ENST00000250113.7
ENST00000571597.1 |
FXR2
|
fragile X mental retardation, autosomal homolog 2 |
| chr19_+_51153045 | 0.33 |
ENST00000458538.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr6_+_43140095 | 0.33 |
ENST00000457278.2
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor) |
| chr11_+_47236489 | 0.33 |
ENST00000256996.4
ENST00000378603.3 ENST00000378600.3 ENST00000378601.3 |
DDB2
|
damage-specific DNA binding protein 2, 48kDa |
| chr22_+_38004723 | 0.33 |
ENST00000381756.5
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr2_+_217498105 | 0.33 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr10_-_99094458 | 0.33 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2 |
| chr14_-_69262789 | 0.33 |
ENST00000557022.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr17_+_7155343 | 0.33 |
ENST00000573513.1
ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr20_-_30458019 | 0.33 |
ENST00000486996.1
ENST00000398084.2 |
DUSP15
|
dual specificity phosphatase 15 |
| chr16_-_31021921 | 0.33 |
ENST00000215095.5
|
STX1B
|
syntaxin 1B |
| chr22_+_41865109 | 0.33 |
ENST00000216254.4
ENST00000396512.3 |
ACO2
|
aconitase 2, mitochondrial |
| chr11_+_117070037 | 0.33 |
ENST00000392951.4
ENST00000525531.1 ENST00000278968.6 |
TAGLN
|
transgelin |
| chr16_-_4665023 | 0.32 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr22_-_21213029 | 0.32 |
ENST00000572273.1
ENST00000255882.6 |
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha |
| chr10_+_120789223 | 0.32 |
ENST00000425699.1
|
NANOS1
|
nanos homolog 1 (Drosophila) |
| chr21_-_43916433 | 0.32 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr17_-_42402138 | 0.32 |
ENST00000592857.1
ENST00000586016.1 ENST00000590194.1 ENST00000377095.5 ENST00000588049.1 ENST00000586633.1 ENST00000537904.2 ENST00000585636.1 ENST00000585523.1 ENST00000225308.8 |
SLC25A39
|
solute carrier family 25, member 39 |
| chr1_+_6051526 | 0.32 |
ENST00000378111.1
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr3_-_178789220 | 0.32 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr12_+_70760056 | 0.32 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr20_-_25038804 | 0.31 |
ENST00000323482.4
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1 |
| chr2_-_27603582 | 0.31 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr7_+_4721885 | 0.31 |
ENST00000328914.4
|
FOXK1
|
forkhead box K1 |
| chr20_+_388056 | 0.31 |
ENST00000411647.1
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr16_+_57680043 | 0.31 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr22_-_46283597 | 0.31 |
ENST00000451118.1
|
WI2-85898F10.1
|
WI2-85898F10.1 |
| chr1_+_43148059 | 0.31 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr10_+_103825080 | 0.31 |
ENST00000299238.5
|
HPS6
|
Hermansky-Pudlak syndrome 6 |
| chr11_+_64053005 | 0.31 |
ENST00000538032.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr19_-_16738984 | 0.31 |
ENST00000600060.1
ENST00000263390.3 |
MED26
|
mediator complex subunit 26 |
| chr4_+_141677577 | 0.31 |
ENST00000609937.1
|
RP11-102N12.3
|
RP11-102N12.3 |
| chr22_-_31741757 | 0.30 |
ENST00000215919.3
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr10_-_95242044 | 0.30 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr19_-_55770311 | 0.30 |
ENST00000412770.2
|
PPP6R1
|
protein phosphatase 6, regulatory subunit 1 |
| chr1_+_110577229 | 0.30 |
ENST00000369795.3
ENST00000369794.2 |
STRIP1
|
striatin interacting protein 1 |
| chr20_+_32254286 | 0.30 |
ENST00000330271.4
|
ACTL10
|
actin-like 10 |
| chr11_+_47291193 | 0.30 |
ENST00000428807.1
ENST00000402799.1 ENST00000406482.1 ENST00000349238.3 ENST00000311027.5 ENST00000407859.3 ENST00000395344.3 ENST00000444117.1 |
MADD
|
MAP-kinase activating death domain |
| chr16_-_122619 | 0.30 |
ENST00000262316.6
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila) |
| chr19_+_507299 | 0.30 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr19_-_18717627 | 0.30 |
ENST00000392386.3
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr9_-_130639997 | 0.30 |
ENST00000373176.1
|
AK1
|
adenylate kinase 1 |
| chr10_+_88718397 | 0.30 |
ENST00000372017.3
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr22_-_20104700 | 0.29 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr6_-_42946888 | 0.29 |
ENST00000244546.4
|
PEX6
|
peroxisomal biogenesis factor 6 |
| chr14_+_74004051 | 0.29 |
ENST00000557556.1
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr10_+_99079008 | 0.29 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr19_+_56146352 | 0.29 |
ENST00000592881.1
|
ZNF580
|
zinc finger protein 580 |
| chr20_-_48532046 | 0.28 |
ENST00000543716.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr8_+_22250334 | 0.28 |
ENST00000520832.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr6_+_155316390 | 0.28 |
ENST00000545347.1
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr19_+_54058073 | 0.28 |
ENST00000505949.1
ENST00000513265.1 |
ZNF331
|
zinc finger protein 331 |
| chr11_-_65686586 | 0.28 |
ENST00000438576.2
|
C11orf68
|
chromosome 11 open reading frame 68 |
| chr11_-_65686496 | 0.28 |
ENST00000449692.3
|
C11orf68
|
chromosome 11 open reading frame 68 |
| chr9_+_140149625 | 0.28 |
ENST00000343053.4
|
NELFB
|
negative elongation factor complex member B |
| chr19_-_55677999 | 0.28 |
ENST00000532817.1
ENST00000527223.2 ENST00000391720.4 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr14_-_69262916 | 0.28 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr5_-_149682447 | 0.27 |
ENST00000328668.7
|
ARSI
|
arylsulfatase family, member I |
| chr20_+_34287194 | 0.27 |
ENST00000374078.1
ENST00000374077.3 |
ROMO1
|
reactive oxygen species modulator 1 |
| chr22_-_30783075 | 0.27 |
ENST00000215798.6
|
RNF215
|
ring finger protein 215 |
| chr14_-_91282821 | 0.27 |
ENST00000553948.1
|
TTC7B
|
tetratricopeptide repeat domain 7B |
| chr13_+_49794474 | 0.27 |
ENST00000218721.1
ENST00000398307.1 |
MLNR
|
motilin receptor |
| chr17_+_7835419 | 0.27 |
ENST00000576538.1
ENST00000380262.3 ENST00000563694.1 ENST00000380255.3 ENST00000570782.1 |
CNTROB
|
centrobin, centrosomal BRCA2 interacting protein |
| chr22_-_32058166 | 0.27 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr16_-_787728 | 0.27 |
ENST00000567403.1
ENST00000562421.1 |
NARFL
|
nuclear prelamin A recognition factor-like |
| chr14_-_73493784 | 0.27 |
ENST00000553891.1
|
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr12_-_50222187 | 0.27 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr20_+_42295745 | 0.27 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr17_-_42298331 | 0.27 |
ENST00000343638.5
|
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr2_-_25475120 | 0.27 |
ENST00000380746.4
ENST00000402667.1 |
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr11_+_118938485 | 0.27 |
ENST00000300793.6
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae) |
| chr20_-_48532019 | 0.27 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr5_+_92919043 | 0.27 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr5_+_175976324 | 0.27 |
ENST00000261944.5
|
CDHR2
|
cadherin-related family member 2 |
| chr1_-_233431458 | 0.27 |
ENST00000258229.9
ENST00000430153.1 |
PCNXL2
|
pecanex-like 2 (Drosophila) |
| chr21_-_43916296 | 0.27 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr11_+_71939512 | 0.26 |
ENST00000540329.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr19_+_39926791 | 0.26 |
ENST00000594990.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr17_-_1395954 | 0.26 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr11_+_111411384 | 0.26 |
ENST00000375615.3
ENST00000525126.1 ENST00000436913.2 ENST00000533265.1 |
LAYN
|
layilin |
| chr4_+_56814968 | 0.26 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr16_+_31044812 | 0.26 |
ENST00000313843.3
|
STX4
|
syntaxin 4 |
| chr22_+_22673051 | 0.26 |
ENST00000390289.2
|
IGLV5-52
|
immunoglobulin lambda variable 5-52 |
| chr19_+_52264449 | 0.26 |
ENST00000599326.1
ENST00000598953.1 |
FPR2
|
formyl peptide receptor 2 |
| chr3_+_160117062 | 0.26 |
ENST00000497311.1
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr22_+_38005033 | 0.26 |
ENST00000447515.1
ENST00000406772.1 ENST00000431745.1 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr12_+_6930964 | 0.26 |
ENST00000382315.3
|
GPR162
|
G protein-coupled receptor 162 |
| chr9_+_130186653 | 0.26 |
ENST00000342483.5
ENST00000543471.1 |
ZNF79
|
zinc finger protein 79 |
| chr6_-_42946947 | 0.26 |
ENST00000304611.8
|
PEX6
|
peroxisomal biogenesis factor 6 |
| chr19_+_41222998 | 0.26 |
ENST00000263370.2
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr12_-_46121554 | 0.26 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr17_-_7155274 | 0.26 |
ENST00000318988.6
ENST00000575783.1 ENST00000573600.1 |
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr1_+_229385383 | 0.26 |
ENST00000323223.2
|
TMEM78
|
transmembrane protein 78 |
| chr10_-_103880209 | 0.25 |
ENST00000425280.1
|
LDB1
|
LIM domain binding 1 |
| chr19_+_41699135 | 0.25 |
ENST00000542619.1
ENST00000600561.1 |
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr19_+_18544045 | 0.25 |
ENST00000599699.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr17_-_1553346 | 0.25 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr19_-_19051993 | 0.25 |
ENST00000594794.1
ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr20_-_4229721 | 0.25 |
ENST00000379453.4
|
ADRA1D
|
adrenoceptor alpha 1D |
| chr19_-_3971050 | 0.25 |
ENST00000545797.2
ENST00000596311.1 |
DAPK3
|
death-associated protein kinase 3 |
| chr16_-_69364467 | 0.25 |
ENST00000288022.1
|
PDF
|
peptide deformylase (mitochondrial) |
| chr17_-_43339474 | 0.25 |
ENST00000331780.4
|
SPATA32
|
spermatogenesis associated 32 |
| chr18_+_8705588 | 0.25 |
ENST00000306329.11
|
SOGA2
|
SOGA family member 2 |
| chr19_-_45735138 | 0.25 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr10_-_5931794 | 0.25 |
ENST00000380092.4
ENST00000380094.5 ENST00000191063.8 |
ANKRD16
|
ankyrin repeat domain 16 |
| chr17_-_79604075 | 0.25 |
ENST00000374747.5
ENST00000539314.1 ENST00000331134.6 |
NPLOC4
|
nuclear protein localization 4 homolog (S. cerevisiae) |
| chr14_-_61190754 | 0.25 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr20_-_32274179 | 0.25 |
ENST00000343380.5
|
E2F1
|
E2F transcription factor 1 |
| chr6_+_30881982 | 0.25 |
ENST00000321897.5
ENST00000416670.2 ENST00000542001.1 ENST00000428017.1 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr1_-_23520755 | 0.25 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr12_+_26348246 | 0.25 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr8_-_38325219 | 0.24 |
ENST00000533668.1
ENST00000413133.2 ENST00000397108.4 ENST00000526742.1 ENST00000525001.1 ENST00000425967.3 ENST00000529552.1 ENST00000397113.2 |
FGFR1
|
fibroblast growth factor receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.2 | 1.0 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.2 | 0.6 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 | 0.8 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.2 | 0.6 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.2 | 0.7 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.2 | 0.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.7 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.1 | 0.4 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.1 | 1.3 | GO:1904636 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.4 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 0.4 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.1 | 0.4 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.6 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.4 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 0.4 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 | 0.3 | GO:0060532 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.1 | 0.3 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.1 | 0.4 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.1 | 0.4 | GO:0019230 | proprioception(GO:0019230) |
| 0.1 | 1.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.4 | GO:2000506 | negative regulation of energy homeostasis(GO:2000506) |
| 0.1 | 0.5 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.3 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.1 | 0.5 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 0.3 | GO:1902568 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.3 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 0.2 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 0.2 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.1 | 0.2 | GO:1902811 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 | 0.2 | GO:0071315 | cellular response to morphine(GO:0071315) cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.1 | 0.2 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.3 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.2 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 0.3 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.1 | 0.4 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 | 0.2 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.2 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.1 | 0.2 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 0.4 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.3 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.1 | 0.4 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.3 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 1.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.2 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.1 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.3 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.5 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.3 | GO:0019541 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.1 | 0.2 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0033076 | isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.4 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.5 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.4 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.0 | 0.2 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.5 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.1 | GO:1904779 | regulation of protein localization to centrosome(GO:1904779) |
| 0.0 | 0.3 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 0.2 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.2 | GO:0070384 | growth plate cartilage chondrocyte growth(GO:0003430) Harderian gland development(GO:0070384) |
| 0.0 | 0.3 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0036034 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.0 | 0.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.7 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.2 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.5 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.3 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.3 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.0 | 0.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.2 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.3 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.2 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.3 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.2 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.4 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 1.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.2 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.3 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 1.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 1.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.3 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.4 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.5 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.5 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.5 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.1 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:0008052 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.3 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.1 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.1 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.2 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0035898 | parathyroid hormone secretion(GO:0035898) post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.2 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) histone H2A K63-linked ubiquitination(GO:0070535) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.3 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.0 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.2 | GO:0044146 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:0002911 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.1 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.3 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.0 | GO:1903527 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.3 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.3 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.6 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.6 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 0.1 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 1.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) cellular polysaccharide metabolic process(GO:0044264) |
| 0.0 | 0.2 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.1 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.2 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.6 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.2 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) regulation of GTP binding(GO:1904424) |
| 0.0 | 0.4 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.2 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 0.8 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.1 | 1.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.4 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.1 | 0.5 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.5 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.2 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.8 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.4 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.3 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.3 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.1 | 0.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.2 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.2 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.6 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 1.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 2.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.6 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.2 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.3 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 0.7 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.2 | 0.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 1.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.4 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.1 | 0.3 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.3 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.1 | 0.4 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.6 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.2 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.1 | 0.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 1.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.3 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 0.1 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 0.3 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.2 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.5 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 0.2 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.2 | GO:0031177 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.4 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 0.3 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.2 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 0.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 1.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.3 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.2 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.2 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 1.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.4 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.6 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0005165 | neurotrophin receptor binding(GO:0005165) neurotrophin TRK receptor binding(GO:0005167) neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) caspase binding(GO:0089720) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 1.0 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 2.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 2.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |