Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
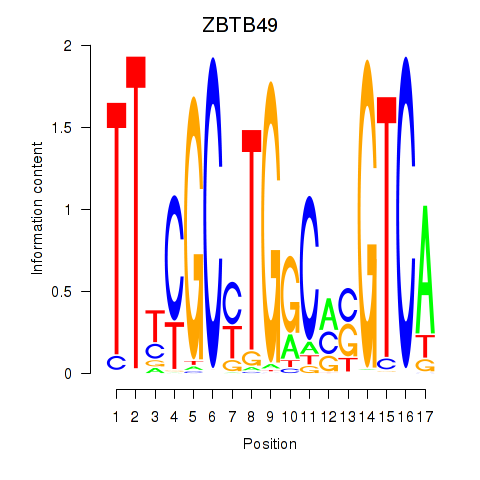
Results for ZBTB49
Z-value: 0.12
Transcription factors associated with ZBTB49
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB49
|
ENSG00000168826.11 | zinc finger and BTB domain containing 49 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB49 | hg19_v2_chr4_+_4291924_4292011 | -0.77 | 7.5e-02 | Click! |
Activity profile of ZBTB49 motif
Sorted Z-values of ZBTB49 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_53525573 | 0.14 |
ENST00000503051.1
|
USP46-AS1
|
USP46 antisense RNA 1 |
| chr15_+_75639296 | 0.11 |
ENST00000564500.1
ENST00000355059.4 ENST00000566752.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr16_+_811073 | 0.11 |
ENST00000382862.3
ENST00000563651.1 |
MSLN
|
mesothelin |
| chr3_-_196242233 | 0.10 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr20_+_1115821 | 0.09 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr16_+_810728 | 0.08 |
ENST00000563941.1
ENST00000545450.2 ENST00000566549.1 |
MSLN
|
mesothelin |
| chr2_+_47799601 | 0.08 |
ENST00000601243.1
|
AC138655.1
|
CDNA: FLJ23120 fis, clone LNG07989; HCG1987724; Uncharacterized protein |
| chr10_-_90751038 | 0.08 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr1_-_151319654 | 0.08 |
ENST00000430227.1
ENST00000412774.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr20_+_5731083 | 0.08 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr2_+_30670127 | 0.06 |
ENST00000540623.1
ENST00000476038.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr8_+_99129513 | 0.06 |
ENST00000522319.1
ENST00000401707.2 |
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr2_+_95873257 | 0.05 |
ENST00000425953.1
|
AC092835.2
|
AC092835.2 |
| chr5_+_98264867 | 0.05 |
ENST00000513175.1
|
CTD-2007H13.3
|
CTD-2007H13.3 |
| chr1_-_151319318 | 0.05 |
ENST00000436271.1
ENST00000450506.1 ENST00000422595.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr16_-_28621353 | 0.05 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr11_+_108094786 | 0.05 |
ENST00000601453.1
|
ATM
|
ataxia telangiectasia mutated |
| chr16_+_2533020 | 0.04 |
ENST00000562105.1
|
TBC1D24
|
TBC1 domain family, member 24 |
| chr6_-_109776901 | 0.04 |
ENST00000431946.1
|
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr5_+_140474181 | 0.04 |
ENST00000194155.4
|
PCDHB2
|
protocadherin beta 2 |
| chr7_+_148936732 | 0.04 |
ENST00000335870.2
|
ZNF212
|
zinc finger protein 212 |
| chr19_-_6481776 | 0.04 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr17_-_80009650 | 0.03 |
ENST00000310496.4
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_-_109777128 | 0.03 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr18_+_43753500 | 0.03 |
ENST00000587591.1
ENST00000588730.1 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr8_+_24151553 | 0.03 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr8_+_24151620 | 0.03 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr19_-_51611623 | 0.03 |
ENST00000421832.2
|
CTU1
|
cytosolic thiouridylase subunit 1 |
| chr16_-_18937726 | 0.03 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr22_-_20256054 | 0.03 |
ENST00000043402.7
|
RTN4R
|
reticulon 4 receptor |
| chr3_+_39424828 | 0.03 |
ENST00000273158.4
ENST00000431510.1 |
SLC25A38
|
solute carrier family 25, member 38 |
| chr19_+_50145328 | 0.03 |
ENST00000360565.3
|
SCAF1
|
SR-related CTD-associated factor 1 |
| chrX_+_101854426 | 0.03 |
ENST00000536530.1
ENST00000473968.1 ENST00000604957.1 ENST00000477663.1 ENST00000479502.1 |
ARMCX5
|
armadillo repeat containing, X-linked 5 |
| chr22_-_20255212 | 0.03 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr2_-_85555086 | 0.03 |
ENST00000444342.2
ENST00000409232.3 ENST00000409015.1 |
TGOLN2
|
trans-golgi network protein 2 |
| chr6_-_159240415 | 0.03 |
ENST00000367075.3
|
EZR
|
ezrin |
| chr11_-_2950642 | 0.03 |
ENST00000314222.4
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2 |
| chr3_-_196756646 | 0.03 |
ENST00000439320.1
ENST00000296351.4 ENST00000296350.5 |
MFI2
|
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
| chr2_+_234160217 | 0.02 |
ENST00000392017.4
ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr1_+_240255166 | 0.02 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr22_-_29196511 | 0.02 |
ENST00000344347.5
|
XBP1
|
X-box binding protein 1 |
| chr17_-_80231557 | 0.02 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
| chr12_-_120241187 | 0.02 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chrX_+_16185604 | 0.02 |
ENST00000400004.2
|
MAGEB17
|
melanoma antigen family B, 17 |
| chr2_+_27237615 | 0.02 |
ENST00000458529.1
ENST00000402218.1 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr19_+_18669809 | 0.02 |
ENST00000602094.1
|
KXD1
|
KxDL motif containing 1 |
| chr8_-_42396632 | 0.02 |
ENST00000520179.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr15_+_75639372 | 0.02 |
ENST00000566313.1
ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr12_+_53835508 | 0.02 |
ENST00000551003.1
ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13
PCBP2
|
proline rich 13 poly(rC) binding protein 2 |
| chr16_+_30406721 | 0.02 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chr1_+_46806452 | 0.02 |
ENST00000536062.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chrX_+_101854096 | 0.02 |
ENST00000246174.2
ENST00000537008.1 ENST00000541409.1 |
ARMCX5
|
armadillo repeat containing, X-linked 5 |
| chr18_-_11148587 | 0.02 |
ENST00000302079.6
ENST00000580640.1 ENST00000503781.3 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr3_+_52232102 | 0.02 |
ENST00000469224.1
ENST00000394965.2 ENST00000310271.2 ENST00000484952.1 |
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr6_+_53659746 | 0.02 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chrX_+_9431324 | 0.02 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr8_+_58907104 | 0.01 |
ENST00000361488.3
|
FAM110B
|
family with sequence similarity 110, member B |
| chr12_+_53835539 | 0.01 |
ENST00000547368.1
ENST00000379786.4 ENST00000551945.1 |
PRR13
|
proline rich 13 |
| chr2_+_234160340 | 0.01 |
ENST00000417017.1
ENST00000392020.4 ENST00000392018.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr8_-_11996586 | 0.01 |
ENST00000333796.3
|
USP17L2
|
ubiquitin specific peptidase 17-like family member 2 |
| chr22_+_42017459 | 0.01 |
ENST00000405878.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr1_-_151319283 | 0.01 |
ENST00000392746.3
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr2_+_30670209 | 0.01 |
ENST00000497423.1
ENST00000476535.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr5_-_146833222 | 0.01 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr8_-_59572093 | 0.01 |
ENST00000427130.2
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr20_-_25320367 | 0.01 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chr3_-_49055991 | 0.01 |
ENST00000441576.2
ENST00000420952.2 ENST00000341949.4 ENST00000395462.4 |
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr22_-_42017021 | 0.01 |
ENST00000263256.6
|
DESI1
|
desumoylating isopeptidase 1 |
| chr9_-_7800067 | 0.01 |
ENST00000358227.4
|
TMEM261
|
transmembrane protein 261 |
| chr8_-_82024290 | 0.01 |
ENST00000220597.4
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr15_-_55488817 | 0.01 |
ENST00000569386.1
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr22_-_19166343 | 0.01 |
ENST00000215882.5
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr2_+_30670077 | 0.01 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr1_+_46806837 | 0.01 |
ENST00000537428.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chr14_+_35591928 | 0.01 |
ENST00000605870.1
ENST00000557404.3 |
KIAA0391
|
KIAA0391 |
| chr1_+_26869597 | 0.01 |
ENST00000530003.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr8_+_42396712 | 0.01 |
ENST00000518574.1
ENST00000417410.2 ENST00000414154.2 |
SMIM19
|
small integral membrane protein 19 |
| chr20_-_33735070 | 0.01 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr9_-_116102530 | 0.01 |
ENST00000374195.3
ENST00000341761.4 |
WDR31
|
WD repeat domain 31 |
| chr10_+_88428206 | 0.01 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chr14_+_77924204 | 0.01 |
ENST00000555133.1
|
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
| chr16_+_69166418 | 0.01 |
ENST00000314423.7
ENST00000562237.1 ENST00000567460.1 ENST00000566227.1 ENST00000352319.4 ENST00000563094.1 |
CIRH1A
|
cirrhosis, autosomal recessive 1A (cirhin) |
| chr16_+_28722809 | 0.01 |
ENST00000566866.1
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C |
| chr1_+_182808474 | 0.01 |
ENST00000367549.3
|
DHX9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr17_+_80009741 | 0.01 |
ENST00000578552.1
ENST00000320548.4 ENST00000581578.1 ENST00000583885.1 ENST00000583641.1 ENST00000581418.1 ENST00000355130.2 ENST00000306823.6 ENST00000392358.2 |
GPS1
|
G protein pathway suppressor 1 |
| chr16_+_30406423 | 0.01 |
ENST00000524644.1
|
ZNF48
|
zinc finger protein 48 |
| chr3_+_196594727 | 0.01 |
ENST00000445299.2
ENST00000323460.5 ENST00000419026.1 |
SENP5
|
SUMO1/sentrin specific peptidase 5 |
| chr9_+_139553306 | 0.01 |
ENST00000371699.1
|
EGFL7
|
EGF-like-domain, multiple 7 |
| chr16_-_28415162 | 0.01 |
ENST00000398944.3
ENST00000398943.3 ENST00000380876.4 |
EIF3CL
|
eukaryotic translation initiation factor 3, subunit C-like |
| chr6_-_43655511 | 0.01 |
ENST00000372133.3
ENST00000372116.1 ENST00000427312.1 |
MRPS18A
|
mitochondrial ribosomal protein S18A |
| chr3_-_184971817 | 0.01 |
ENST00000440662.1
ENST00000456310.1 |
EHHADH
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr12_+_53835425 | 0.01 |
ENST00000549924.1
|
PRR13
|
proline rich 13 |
| chr16_+_28722684 | 0.00 |
ENST00000331666.6
ENST00000395587.1 ENST00000569690.1 ENST00000564243.1 |
EIF3C
|
eukaryotic translation initiation factor 3, subunit C |
| chr1_+_8021954 | 0.00 |
ENST00000377491.1
ENST00000377488.1 |
PARK7
|
parkinson protein 7 |
| chr2_+_27498331 | 0.00 |
ENST00000402462.1
ENST00000404433.1 ENST00000406962.1 |
DNAJC5G
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma |
| chr14_-_23446900 | 0.00 |
ENST00000556731.1
|
AJUBA
|
ajuba LIM protein |
| chr12_+_53835383 | 0.00 |
ENST00000429243.2
|
PRR13
|
proline rich 13 |
| chr7_-_44613494 | 0.00 |
ENST00000431640.1
ENST00000258772.5 |
DDX56
|
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr16_-_18937480 | 0.00 |
ENST00000532700.2
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr2_-_85555355 | 0.00 |
ENST00000282120.2
ENST00000398263.2 |
TGOLN2
|
trans-golgi network protein 2 |
| chr14_+_79745682 | 0.00 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr8_+_124968225 | 0.00 |
ENST00000399018.1
|
FER1L6
|
fer-1-like 6 (C. elegans) |
| chr4_-_176828307 | 0.00 |
ENST00000513365.1
ENST00000513667.1 ENST00000503563.1 |
GPM6A
|
glycoprotein M6A |
| chr19_-_4182530 | 0.00 |
ENST00000601571.1
ENST00000601488.1 ENST00000305232.6 ENST00000381935.3 ENST00000337491.2 |
SIRT6
|
sirtuin 6 |
| chr22_-_29196546 | 0.00 |
ENST00000403532.3
ENST00000216037.6 |
XBP1
|
X-box binding protein 1 |
| chr22_+_21996549 | 0.00 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr22_-_37915247 | 0.00 |
ENST00000251973.5
|
CARD10
|
caspase recruitment domain family, member 10 |
| chr1_+_9294822 | 0.00 |
ENST00000377403.2
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr1_+_8021713 | 0.00 |
ENST00000338639.5
ENST00000493678.1 ENST00000377493.5 |
PARK7
|
parkinson protein 7 |
| chr14_+_79745746 | 0.00 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr11_+_63993738 | 0.00 |
ENST00000441250.2
ENST00000279206.3 |
NUDT22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr9_-_123676827 | 0.00 |
ENST00000546084.1
|
TRAF1
|
TNF receptor-associated factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB49
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |