Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
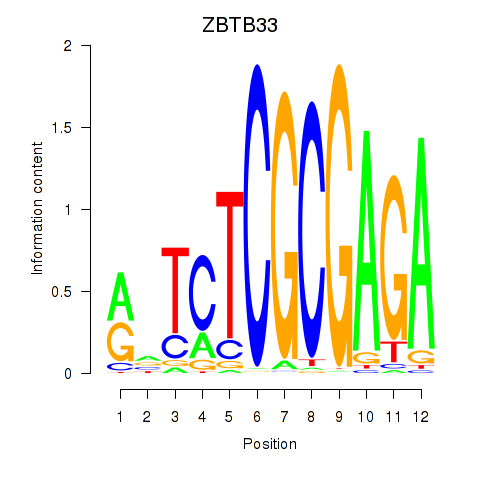
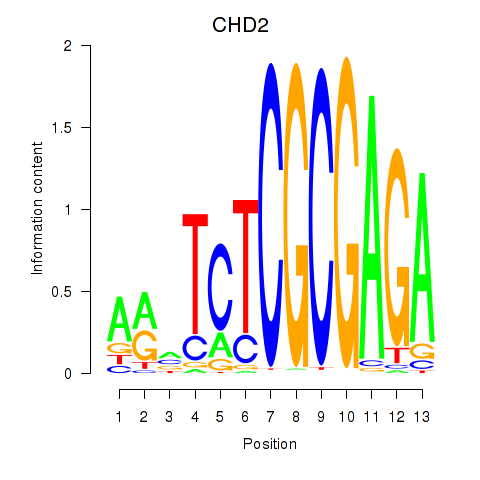
Results for ZBTB33_CHD2
Z-value: 2.21
Transcription factors associated with ZBTB33_CHD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB33
|
ENSG00000177485.6 | zinc finger and BTB domain containing 33 |
|
CHD2
|
ENSG00000173575.14 | chromodomain helicase DNA binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CHD2 | hg19_v2_chr15_+_93447675_93447703 | 0.88 | 2.1e-02 | Click! |
| ZBTB33 | hg19_v2_chrX_+_119384607_119384720 | 0.86 | 2.7e-02 | Click! |
Activity profile of ZBTB33_CHD2 motif
Sorted Z-values of ZBTB33_CHD2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_78730531 | 2.66 |
ENST00000258886.8
|
IREB2
|
iron-responsive element binding protein 2 |
| chr17_+_53046096 | 2.29 |
ENST00000376352.2
ENST00000299341.4 ENST00000405898.1 ENST00000434978.2 ENST00000398391.2 |
STXBP4
|
syntaxin binding protein 4 |
| chr12_-_29534074 | 1.89 |
ENST00000546839.1
ENST00000360150.4 ENST00000552155.1 ENST00000550353.1 ENST00000548441.1 ENST00000552132.1 |
ERGIC2
|
ERGIC and golgi 2 |
| chr3_-_145878954 | 1.76 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr4_-_103749105 | 1.74 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_-_100016492 | 1.74 |
ENST00000369217.4
ENST00000369220.4 ENST00000482541.2 |
CCNC
|
cyclin C |
| chr2_-_136743039 | 1.64 |
ENST00000537273.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr12_-_27091183 | 1.63 |
ENST00000544548.1
ENST00000261191.7 ENST00000537336.1 |
ASUN
|
asunder spermatogenesis regulator |
| chr1_+_111682827 | 1.58 |
ENST00000357172.4
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr5_+_169011033 | 1.57 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr4_-_103749313 | 1.57 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_-_100016527 | 1.55 |
ENST00000523985.1
ENST00000518714.1 ENST00000520371.1 |
CCNC
|
cyclin C |
| chr12_-_27090896 | 1.54 |
ENST00000539625.1
ENST00000538727.1 |
ASUN
|
asunder spermatogenesis regulator |
| chr15_+_78730622 | 1.52 |
ENST00000560440.1
|
IREB2
|
iron-responsive element binding protein 2 |
| chr6_-_100016678 | 1.47 |
ENST00000523799.1
ENST00000520429.1 |
CCNC
|
cyclin C |
| chr18_-_59854203 | 1.36 |
ENST00000589339.1
ENST00000357637.5 ENST00000585458.1 ENST00000400334.3 ENST00000587134.1 ENST00000585923.1 ENST00000590765.1 ENST00000589720.1 ENST00000588571.1 ENST00000585344.1 |
PIGN
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr1_-_207224307 | 1.36 |
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase |
| chr10_-_127408011 | 1.30 |
ENST00000531977.1
ENST00000527483.1 ENST00000525909.1 ENST00000528844.1 ENST00000423178.2 |
RP11-383C5.4
|
RP11-383C5.4 |
| chr16_-_84150410 | 1.27 |
ENST00000569907.1
|
MBTPS1
|
membrane-bound transcription factor peptidase, site 1 |
| chr4_+_17812525 | 1.26 |
ENST00000251496.2
|
NCAPG
|
non-SMC condensin I complex, subunit G |
| chr6_-_97731019 | 1.26 |
ENST00000275053.4
|
MMS22L
|
MMS22-like, DNA repair protein |
| chr4_+_26321284 | 1.25 |
ENST00000506956.1
ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr11_+_94227129 | 1.25 |
ENST00000540349.1
ENST00000535502.1 ENST00000545130.1 ENST00000544253.1 ENST00000541144.1 |
ANKRD49
|
ankyrin repeat domain 49 |
| chr10_-_112064665 | 1.24 |
ENST00000369603.5
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr12_+_102514019 | 1.22 |
ENST00000537257.1
ENST00000358383.5 ENST00000392911.2 |
PARPBP
|
PARP1 binding protein |
| chr1_-_78148324 | 1.21 |
ENST00000370801.3
ENST00000433749.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr5_-_89705537 | 1.17 |
ENST00000522864.1
ENST00000522083.1 ENST00000522565.1 ENST00000522842.1 ENST00000283122.3 |
CETN3
|
centrin, EF-hand protein, 3 |
| chr3_-_160117301 | 1.12 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr1_+_111992064 | 1.12 |
ENST00000483994.1
|
ATP5F1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 |
| chr13_-_21750659 | 1.11 |
ENST00000400018.3
ENST00000314759.5 |
SKA3
|
spindle and kinetochore associated complex subunit 3 |
| chr3_+_108308559 | 1.11 |
ENST00000486815.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr3_-_160117035 | 1.10 |
ENST00000489004.1
ENST00000496589.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr1_-_111682662 | 1.09 |
ENST00000286692.4
|
DRAM2
|
DNA-damage regulated autophagy modulator 2 |
| chr12_+_102513950 | 1.08 |
ENST00000378128.3
ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP
|
PARP1 binding protein |
| chr20_-_34330129 | 1.04 |
ENST00000397370.3
ENST00000528062.3 ENST00000407261.4 ENST00000374038.3 ENST00000361162.6 |
RBM39
|
RNA binding motif protein 39 |
| chr12_+_123237321 | 1.03 |
ENST00000280557.6
ENST00000455982.2 |
DENR
|
density-regulated protein |
| chr10_+_127408263 | 1.03 |
ENST00000337623.3
|
C10orf137
|
erythroid differentiation regulatory factor 1 |
| chr3_+_93781728 | 1.03 |
ENST00000314622.4
|
NSUN3
|
NOP2/Sun domain family, member 3 |
| chr1_-_59165763 | 1.02 |
ENST00000472487.1
|
MYSM1
|
Myb-like, SWIRM and MPN domains 1 |
| chr8_-_42698292 | 1.02 |
ENST00000529779.1
|
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr7_+_35840819 | 1.01 |
ENST00000399035.3
|
SEPT7
|
septin 7 |
| chr1_-_111682813 | 1.01 |
ENST00000539140.1
|
DRAM2
|
DNA-damage regulated autophagy modulator 2 |
| chr10_+_96305610 | 0.99 |
ENST00000371332.4
ENST00000239026.6 |
HELLS
|
helicase, lymphoid-specific |
| chr3_+_108308845 | 0.99 |
ENST00000479138.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr13_+_42846272 | 0.96 |
ENST00000025301.2
|
AKAP11
|
A kinase (PRKA) anchor protein 11 |
| chr4_+_15683404 | 0.95 |
ENST00000422728.2
|
FAM200B
|
family with sequence similarity 200, member B |
| chr2_+_201676908 | 0.94 |
ENST00000409226.1
ENST00000452790.2 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr21_+_47013566 | 0.94 |
ENST00000441095.1
ENST00000424569.1 |
AL133493.2
|
AL133493.2 |
| chr12_-_80328949 | 0.93 |
ENST00000450142.2
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr10_+_127408110 | 0.88 |
ENST00000356792.4
|
C10orf137
|
erythroid differentiation regulatory factor 1 |
| chr5_+_56469843 | 0.87 |
ENST00000514387.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr17_+_28443819 | 0.87 |
ENST00000479218.2
|
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr8_+_38088861 | 0.86 |
ENST00000397166.2
ENST00000533100.1 |
DDHD2
|
DDHD domain containing 2 |
| chr9_+_26956474 | 0.85 |
ENST00000429045.2
|
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr11_-_93474645 | 0.84 |
ENST00000532455.1
|
TAF1D
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa |
| chr3_-_108308241 | 0.84 |
ENST00000295746.8
|
KIAA1524
|
KIAA1524 |
| chr7_+_35840542 | 0.83 |
ENST00000435235.1
ENST00000399034.2 ENST00000350320.6 ENST00000469679.2 |
SEPT7
|
septin 7 |
| chr13_-_22178284 | 0.83 |
ENST00000468222.2
ENST00000382374.4 |
MICU2
|
mitochondrial calcium uptake 2 |
| chr9_+_86595626 | 0.83 |
ENST00000445877.1
ENST00000325875.3 |
RMI1
|
RecQ mediated genome instability 1 |
| chr9_+_26956371 | 0.82 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr8_-_42698433 | 0.82 |
ENST00000345117.2
ENST00000254250.3 |
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr2_-_74405929 | 0.82 |
ENST00000396049.4
|
MOB1A
|
MOB kinase activator 1A |
| chr1_+_174969262 | 0.82 |
ENST00000406752.1
ENST00000405362.1 |
CACYBP
|
calcyclin binding protein |
| chr14_+_64970427 | 0.81 |
ENST00000553583.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr2_+_198318217 | 0.81 |
ENST00000409398.1
|
COQ10B
|
coenzyme Q10 homolog B (S. cerevisiae) |
| chr6_-_7313381 | 0.80 |
ENST00000489567.1
ENST00000479365.1 ENST00000462112.1 ENST00000397511.2 ENST00000534851.1 ENST00000474597.1 ENST00000244763.4 |
SSR1
|
signal sequence receptor, alpha |
| chr10_+_96305535 | 0.80 |
ENST00000419900.1
ENST00000348459.5 ENST00000394045.1 ENST00000394044.1 ENST00000394036.1 |
HELLS
|
helicase, lymphoid-specific |
| chr4_+_25378826 | 0.80 |
ENST00000315368.3
|
ANAPC4
|
anaphase promoting complex subunit 4 |
| chr14_+_74318611 | 0.79 |
ENST00000555976.1
ENST00000267568.4 |
PTGR2
|
prostaglandin reductase 2 |
| chr11_+_107879459 | 0.79 |
ENST00000393094.2
|
CUL5
|
cullin 5 |
| chr4_+_26322409 | 0.78 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr1_-_207226313 | 0.78 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr10_-_7829909 | 0.78 |
ENST00000379562.4
ENST00000543003.1 ENST00000535925.1 |
KIN
|
KIN, antigenic determinant of recA protein homolog (mouse) |
| chr2_-_55844720 | 0.77 |
ENST00000345102.5
ENST00000272313.5 ENST00000407823.3 |
SMEK2
|
SMEK homolog 2, suppressor of mek1 (Dictyostelium) |
| chr2_+_201676256 | 0.77 |
ENST00000452206.1
ENST00000410110.2 ENST00000409600.1 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr4_+_120133791 | 0.76 |
ENST00000274030.6
|
USP53
|
ubiquitin specific peptidase 53 |
| chr15_-_40074996 | 0.75 |
ENST00000350221.3
|
FSIP1
|
fibrous sheath interacting protein 1 |
| chr4_-_103748880 | 0.75 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_+_16884740 | 0.75 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3 |
| chr12_+_27091426 | 0.74 |
ENST00000546072.1
ENST00000327214.5 |
FGFR1OP2
|
FGFR1 oncogene partner 2 |
| chr3_+_160117087 | 0.74 |
ENST00000357388.3
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr6_-_108279369 | 0.73 |
ENST00000369002.4
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr3_-_160116995 | 0.73 |
ENST00000465537.1
ENST00000486856.1 ENST00000468218.1 ENST00000478370.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr2_-_20101701 | 0.73 |
ENST00000402414.1
ENST00000333610.3 |
TTC32
|
tetratricopeptide repeat domain 32 |
| chr2_+_214149113 | 0.73 |
ENST00000331683.5
ENST00000432529.2 ENST00000413312.1 ENST00000272898.7 ENST00000447990.1 |
SPAG16
|
sperm associated antigen 16 |
| chr11_-_94226964 | 0.72 |
ENST00000538923.1
ENST00000540013.1 ENST00000407439.3 ENST00000393241.4 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr1_+_104068562 | 0.72 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr3_+_108308513 | 0.70 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr2_+_207630081 | 0.69 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr2_-_136743169 | 0.68 |
ENST00000264161.4
|
DARS
|
aspartyl-tRNA synthetase |
| chr17_+_65714018 | 0.68 |
ENST00000581106.1
ENST00000535137.1 |
NOL11
|
nucleolar protein 11 |
| chr8_+_33342268 | 0.68 |
ENST00000360128.6
|
MAK16
|
MAK16 homolog (S. cerevisiae) |
| chr2_+_160568978 | 0.67 |
ENST00000409175.1
ENST00000539065.1 ENST00000259050.4 ENST00000421037.1 |
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chr4_+_83821835 | 0.67 |
ENST00000302236.5
|
THAP9
|
THAP domain containing 9 |
| chr9_-_123605177 | 0.67 |
ENST00000373904.5
ENST00000210313.3 |
PSMD5
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
| chr3_+_14219858 | 0.67 |
ENST00000306024.3
|
LSM3
|
LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr5_+_56469939 | 0.67 |
ENST00000506184.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr1_-_24306768 | 0.67 |
ENST00000374453.3
ENST00000453840.3 |
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr21_-_34144157 | 0.66 |
ENST00000331923.4
|
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr12_+_27091316 | 0.66 |
ENST00000229395.3
|
FGFR1OP2
|
FGFR1 oncogene partner 2 |
| chr2_+_99758161 | 0.66 |
ENST00000409684.1
|
C2ORF15
|
Uncharacterized protein C2orf15 |
| chr12_-_80328700 | 0.65 |
ENST00000550107.1
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr17_+_65713925 | 0.65 |
ENST00000253247.4
|
NOL11
|
nucleolar protein 11 |
| chr11_+_11863579 | 0.65 |
ENST00000399455.2
|
USP47
|
ubiquitin specific peptidase 47 |
| chrX_+_13752832 | 0.64 |
ENST00000380550.3
ENST00000398395.3 ENST00000340096.6 ENST00000380567.1 |
OFD1
|
oral-facial-digital syndrome 1 |
| chr20_-_25604811 | 0.64 |
ENST00000304788.3
|
NANP
|
N-acetylneuraminic acid phosphatase |
| chr8_+_38089198 | 0.64 |
ENST00000528358.1
ENST00000529642.1 ENST00000532222.1 ENST00000520272.2 |
DDHD2
|
DDHD domain containing 2 |
| chr17_-_53046058 | 0.63 |
ENST00000571584.1
ENST00000299335.3 |
COX11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr6_+_71122974 | 0.63 |
ENST00000418814.2
|
FAM135A
|
family with sequence similarity 135, member A |
| chr5_-_39074479 | 0.62 |
ENST00000514735.1
ENST00000296782.5 ENST00000357387.3 |
RICTOR
|
RPTOR independent companion of MTOR, complex 2 |
| chr15_+_66797455 | 0.62 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr2_+_149402009 | 0.62 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr12_+_67663056 | 0.62 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr3_-_113464906 | 0.62 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chrY_+_15016725 | 0.61 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr3_+_160117418 | 0.61 |
ENST00000465903.1
ENST00000485645.1 ENST00000360111.2 ENST00000472991.1 ENST00000467468.1 ENST00000469762.1 ENST00000489573.1 ENST00000462787.1 ENST00000490207.1 ENST00000485867.1 |
SMC4
|
structural maintenance of chromosomes 4 |
| chr15_-_66649010 | 0.61 |
ENST00000367709.4
ENST00000261881.4 |
TIPIN
|
TIMELESS interacting protein |
| chr7_-_119547421 | 0.61 |
ENST00000431071.1
ENST00000426413.1 |
RP11-328J2.1
|
RP11-328J2.1 |
| chrX_+_41192595 | 0.60 |
ENST00000399959.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr6_+_71123107 | 0.60 |
ENST00000370479.3
ENST00000505769.1 ENST00000515323.1 ENST00000515280.1 ENST00000507085.1 ENST00000457062.2 ENST00000361499.3 |
FAM135A
|
family with sequence similarity 135, member A |
| chr3_+_182511266 | 0.60 |
ENST00000323116.5
ENST00000493826.1 |
ATP11B
|
ATPase, class VI, type 11B |
| chr10_+_51565108 | 0.60 |
ENST00000438493.1
ENST00000452682.1 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr11_+_11863500 | 0.60 |
ENST00000527733.1
ENST00000539466.1 |
USP47
|
ubiquitin specific peptidase 47 |
| chr2_-_70520539 | 0.60 |
ENST00000482975.2
ENST00000438261.1 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr2_-_136743436 | 0.59 |
ENST00000441323.1
ENST00000449218.1 |
DARS
|
aspartyl-tRNA synthetase |
| chr3_-_93781750 | 0.59 |
ENST00000314636.2
|
DHFRL1
|
dihydrofolate reductase-like 1 |
| chr7_-_129845313 | 0.59 |
ENST00000397622.2
|
TMEM209
|
transmembrane protein 209 |
| chr21_-_30445886 | 0.59 |
ENST00000431234.1
ENST00000540844.1 ENST00000286788.4 |
CCT8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr4_-_103748696 | 0.58 |
ENST00000321805.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr5_-_159846066 | 0.58 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr9_+_88556036 | 0.57 |
ENST00000361671.5
ENST00000416045.1 |
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr2_+_9563769 | 0.57 |
ENST00000475482.1
|
CPSF3
|
cleavage and polyadenylation specific factor 3, 73kDa |
| chr14_+_64970662 | 0.57 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr11_-_27528301 | 0.57 |
ENST00000524596.1
ENST00000278193.2 |
LIN7C
|
lin-7 homolog C (C. elegans) |
| chr5_+_138940742 | 0.57 |
ENST00000398733.3
ENST00000253815.2 ENST00000505007.1 |
UBE2D2
|
ubiquitin-conjugating enzyme E2D 2 |
| chr15_+_66797627 | 0.56 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr6_+_44355257 | 0.56 |
ENST00000371477.3
|
CDC5L
|
cell division cycle 5-like |
| chr7_-_91509986 | 0.56 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr20_-_17949100 | 0.56 |
ENST00000431277.1
|
SNX5
|
sorting nexin 5 |
| chr12_+_28299014 | 0.55 |
ENST00000538586.1
ENST00000536154.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr5_+_56469775 | 0.55 |
ENST00000424459.3
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr2_+_170655789 | 0.55 |
ENST00000409333.1
|
SSB
|
Sjogren syndrome antigen B (autoantigen La) |
| chr11_-_94227029 | 0.55 |
ENST00000323977.3
ENST00000536754.1 ENST00000323929.3 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr1_+_111682058 | 0.54 |
ENST00000545121.1
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr10_+_7830125 | 0.54 |
ENST00000335698.4
ENST00000541227.1 |
ATP5C1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr10_-_5855350 | 0.54 |
ENST00000456041.1
ENST00000380181.3 ENST00000418688.1 ENST00000380132.4 ENST00000609712.1 ENST00000380191.4 |
GDI2
|
GDP dissociation inhibitor 2 |
| chr14_+_45553296 | 0.54 |
ENST00000355765.6
ENST00000553605.1 |
PRPF39
|
pre-mRNA processing factor 39 |
| chr7_-_129845188 | 0.54 |
ENST00000462753.1
ENST00000471077.1 ENST00000473456.1 ENST00000336804.8 |
TMEM209
|
transmembrane protein 209 |
| chr4_+_25378912 | 0.54 |
ENST00000510092.1
ENST00000505991.1 |
ANAPC4
|
anaphase promoting complex subunit 4 |
| chr9_+_98637954 | 0.53 |
ENST00000288985.7
|
ERCC6L2
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like 2 |
| chr11_-_13484713 | 0.53 |
ENST00000526841.1
ENST00000529708.1 ENST00000278174.5 ENST00000528120.1 |
BTBD10
|
BTB (POZ) domain containing 10 |
| chr8_+_104311059 | 0.52 |
ENST00000358755.4
ENST00000523739.1 ENST00000540287.1 |
FZD6
|
frizzled family receptor 6 |
| chr3_+_169684553 | 0.52 |
ENST00000337002.4
ENST00000480708.1 |
SEC62
|
SEC62 homolog (S. cerevisiae) |
| chr16_-_46864955 | 0.52 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr10_-_89577910 | 0.51 |
ENST00000308448.7
ENST00000541004.1 |
ATAD1
|
ATPase family, AAA domain containing 1 |
| chr3_+_42642106 | 0.51 |
ENST00000232978.8
|
NKTR
|
natural killer-tumor recognition sequence |
| chr12_-_49110613 | 0.51 |
ENST00000261900.3
|
CCNT1
|
cyclin T1 |
| chr10_-_101989315 | 0.50 |
ENST00000370397.7
|
CHUK
|
conserved helix-loop-helix ubiquitous kinase |
| chr2_-_25194963 | 0.50 |
ENST00000264711.2
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr10_+_7830092 | 0.50 |
ENST00000356708.7
|
ATP5C1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr5_+_132387633 | 0.50 |
ENST00000304858.2
|
HSPA4
|
heat shock 70kDa protein 4 |
| chr1_+_231473743 | 0.50 |
ENST00000295050.7
|
SPRTN
|
SprT-like N-terminal domain |
| chr10_+_124768482 | 0.50 |
ENST00000368869.4
ENST00000358776.4 |
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain |
| chr3_+_23847432 | 0.48 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr15_-_48470558 | 0.48 |
ENST00000324324.7
|
MYEF2
|
myelin expression factor 2 |
| chr5_+_169010638 | 0.48 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr2_-_200715834 | 0.48 |
ENST00000420128.1
ENST00000416668.1 |
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr1_-_165738072 | 0.48 |
ENST00000481278.1
|
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr20_-_17949143 | 0.48 |
ENST00000419004.1
|
SNX5
|
sorting nexin 5 |
| chr15_-_48470544 | 0.47 |
ENST00000267836.6
|
MYEF2
|
myelin expression factor 2 |
| chr1_+_28052456 | 0.47 |
ENST00000373954.6
ENST00000419687.2 |
FAM76A
|
family with sequence similarity 76, member A |
| chr1_-_27286897 | 0.47 |
ENST00000320567.5
|
C1orf172
|
chromosome 1 open reading frame 172 |
| chr2_-_225450100 | 0.46 |
ENST00000344951.4
|
CUL3
|
cullin 3 |
| chr19_+_36239576 | 0.46 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr5_+_72861560 | 0.46 |
ENST00000296792.4
ENST00000509005.1 ENST00000543251.1 ENST00000508686.1 ENST00000508491.1 |
UTP15
|
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) |
| chr12_-_102513843 | 0.46 |
ENST00000551744.2
ENST00000552283.1 |
NUP37
|
nucleoporin 37kDa |
| chr3_+_169491171 | 0.45 |
ENST00000356716.4
|
MYNN
|
myoneurin |
| chr8_+_103876528 | 0.45 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr7_+_87505544 | 0.45 |
ENST00000265728.1
|
DBF4
|
DBF4 homolog (S. cerevisiae) |
| chr17_-_17184546 | 0.44 |
ENST00000417352.1
|
COPS3
|
COP9 signalosome subunit 3 |
| chr9_-_125667618 | 0.44 |
ENST00000423239.2
|
RC3H2
|
ring finger and CCCH-type domains 2 |
| chr5_-_137878887 | 0.44 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr2_+_170683979 | 0.44 |
ENST00000418381.1
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr3_+_160939050 | 0.43 |
ENST00000493066.1
ENST00000351193.2 ENST00000472947.1 ENST00000463518.1 |
NMD3
|
NMD3 ribosome export adaptor |
| chr18_+_22006646 | 0.43 |
ENST00000585067.1
ENST00000578221.1 |
IMPACT
|
impact RWD domain protein |
| chr12_-_123849374 | 0.43 |
ENST00000602398.1
ENST00000602750.1 |
SBNO1
|
strawberry notch homolog 1 (Drosophila) |
| chr13_-_36920615 | 0.43 |
ENST00000494062.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr14_+_102027688 | 0.43 |
ENST00000510508.4
ENST00000359323.3 |
DIO3
|
deiodinase, iodothyronine, type III |
| chr19_+_34663397 | 0.43 |
ENST00000540746.2
ENST00000544216.3 ENST00000433627.5 |
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae) |
| chr1_+_167906056 | 0.43 |
ENST00000367840.3
|
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr10_-_75385711 | 0.43 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr19_+_37178482 | 0.42 |
ENST00000536254.2
|
ZNF567
|
zinc finger protein 567 |
| chr6_+_47445467 | 0.42 |
ENST00000359314.5
|
CD2AP
|
CD2-associated protein |
| chr2_+_170655322 | 0.42 |
ENST00000260956.4
ENST00000417292.1 |
SSB
|
Sjogren syndrome antigen B (autoantigen La) |
| chr10_+_51565188 | 0.41 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr17_+_54911444 | 0.41 |
ENST00000284061.3
ENST00000572810.1 |
DGKE
|
diacylglycerol kinase, epsilon 64kDa |
| chr1_-_24306798 | 0.41 |
ENST00000374452.5
ENST00000492112.2 ENST00000343255.5 ENST00000344989.6 |
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr4_+_166248775 | 0.41 |
ENST00000261507.6
ENST00000507013.1 ENST00000393766.2 ENST00000504317.1 |
MSMO1
|
methylsterol monooxygenase 1 |
| chr13_+_73356197 | 0.41 |
ENST00000326291.6
|
PIBF1
|
progesterone immunomodulatory binding factor 1 |
| chr14_+_74318513 | 0.40 |
ENST00000555228.1
ENST00000555661.1 |
PTGR2
|
prostaglandin reductase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB33_CHD2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.7 | 2.0 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.5 | 2.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.4 | 1.8 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.4 | 1.8 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.4 | 5.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.3 | 1.4 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.3 | 1.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 1.0 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.3 | 2.6 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.3 | 1.8 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.3 | 0.3 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.3 | 0.8 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.3 | 0.8 | GO:0071264 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.2 | 0.7 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.2 | 0.9 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.2 | 1.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 0.8 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.2 | 0.7 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 0.7 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.2 | 2.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 0.7 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.2 | 0.5 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.2 | 0.9 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.2 | 2.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 1.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 1.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.2 | 0.8 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.6 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 2.3 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 1.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.6 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.7 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.7 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 1.6 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.6 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.1 | 0.6 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.6 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.3 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.3 | GO:0042137 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) sequestering of neurotransmitter(GO:0042137) |
| 0.1 | 0.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.3 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 0.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 5.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 0.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.5 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.5 | GO:0090135 | positive regulation of synapse structural plasticity(GO:0051835) actin filament branching(GO:0090135) |
| 0.1 | 0.4 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.6 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.6 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.6 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.1 | 0.8 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.3 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.7 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.5 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.2 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.9 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 0.4 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 0.5 | GO:1902741 | interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.1 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.2 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.1 | 0.4 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 4.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.4 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.2 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.1 | 1.5 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 0.4 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.1 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.2 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 1.0 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 0.3 | GO:1904806 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.1 | 0.8 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 1.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.7 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.4 | GO:0015853 | adenine transport(GO:0015853) |
| 0.1 | 1.7 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 1.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 2.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.3 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.2 | GO:0048377 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) |
| 0.1 | 0.2 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.2 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.5 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.4 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.3 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 0.2 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 0.6 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 1.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.8 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 2.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 trimethylation(GO:0097198) histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.2 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.7 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.7 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.4 | GO:0007135 | meiosis II(GO:0007135) |
| 0.0 | 0.5 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 4.1 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.4 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 1.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.3 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.4 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.2 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.0 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0061738 | late endosomal microautophagy(GO:0061738) negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.4 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) regulation of endoribonuclease activity(GO:0060699) |
| 0.0 | 0.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.6 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.7 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.2 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.5 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.2 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.2 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 1.4 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.2 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.1 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.3 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.4 | GO:0016241 | regulation of macroautophagy(GO:0016241) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 1.0 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.8 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.4 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.2 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 2.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.0 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.2 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.6 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.3 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.2 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.6 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.0 | GO:0002238 | response to molecule of fungal origin(GO:0002238) cellular response to molecule of fungal origin(GO:0071226) |
| 0.0 | 0.1 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.4 | GO:0014829 | vascular smooth muscle contraction(GO:0014829) |
| 0.0 | 0.5 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.3 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.4 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 0.0 | 0.1 | GO:0031291 | mitotic chromosome movement towards spindle pole(GO:0007079) Ran protein signal transduction(GO:0031291) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.4 | 1.3 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.4 | 1.3 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.4 | 1.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.3 | 1.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.3 | 0.8 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.3 | 2.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 2.8 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 0.7 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 2.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 0.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 0.9 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 3.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.0 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 3.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 4.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 2.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.8 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 0.3 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 0.8 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.1 | 0.8 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 4.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.5 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.2 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.1 | 0.2 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 1.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 2.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.6 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 1.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.9 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.3 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.3 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 2.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.3 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 1.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.5 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.5 | 2.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.5 | 2.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.5 | 2.9 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.5 | 1.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.5 | 1.8 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.4 | 1.8 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.3 | 0.9 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.3 | 0.5 | GO:0016615 | malate dehydrogenase activity(GO:0016615) L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 1.0 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.2 | 2.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.5 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.2 | 0.5 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.2 | 1.2 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.2 | 0.7 | GO:0004803 | transposase activity(GO:0004803) |
| 0.2 | 0.8 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.2 | 0.8 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.5 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 3.0 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 2.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.4 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.1 | 5.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.8 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.3 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 1.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.8 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.4 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 0.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 1.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.5 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 4.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 2.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 2.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.2 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 1.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.2 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.4 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 0.8 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 0.7 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.4 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.3 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.8 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 1.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 1.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 2.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0050610 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.3 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 1.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.0 | GO:0031692 | alpha-1B adrenergic receptor binding(GO:0031692) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 1.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 2.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.6 | GO:0016896 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 1.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 5.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 4.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 3.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 2.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 2.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 4.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 5.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.7 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 2.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.3 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.2 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.0 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 2.3 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.9 | REACTOME G2 M CHECKPOINTS | Genes involved in G2/M Checkpoints |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |