Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
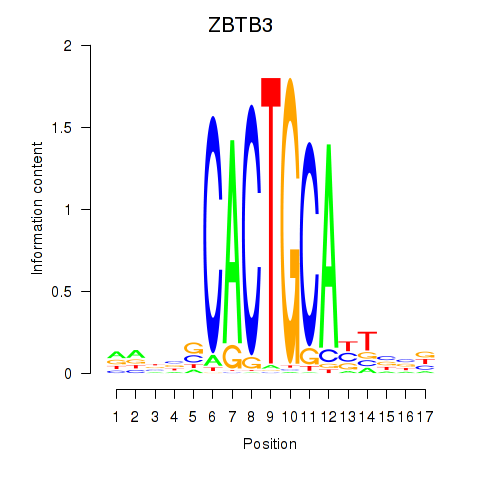
Results for ZBTB3
Z-value: 0.56
Transcription factors associated with ZBTB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB3
|
ENSG00000185670.7 | zinc finger and BTB domain containing 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB3 | hg19_v2_chr11_-_62521614_62521660 | -0.68 | 1.4e-01 | Click! |
Activity profile of ZBTB3 motif
Sorted Z-values of ZBTB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_82782861 | 0.31 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr12_-_10022735 | 0.29 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr16_+_53164956 | 0.28 |
ENST00000563410.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr3_-_195163584 | 0.28 |
ENST00000439666.1
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr3_-_149688502 | 0.24 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chr4_+_48492269 | 0.24 |
ENST00000327939.4
|
ZAR1
|
zygote arrest 1 |
| chr1_-_169555779 | 0.19 |
ENST00000367797.3
ENST00000367796.3 |
F5
|
coagulation factor V (proaccelerin, labile factor) |
| chr11_-_82782952 | 0.19 |
ENST00000534141.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr6_+_30851205 | 0.18 |
ENST00000515881.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr11_+_82783097 | 0.18 |
ENST00000501011.2
ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1
|
RAB30 antisense RNA 1 (head to head) |
| chr10_-_105156198 | 0.18 |
ENST00000369815.1
ENST00000309579.3 ENST00000337003.4 |
USMG5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr3_-_149688896 | 0.16 |
ENST00000239940.7
|
PFN2
|
profilin 2 |
| chr1_+_211433275 | 0.16 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr5_+_115177178 | 0.15 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr1_+_110036674 | 0.15 |
ENST00000393709.3
|
CYB561D1
|
cytochrome b561 family, member D1 |
| chr21_-_48024986 | 0.15 |
ENST00000291700.4
ENST00000367071.4 |
S100B
|
S100 calcium binding protein B |
| chr3_-_149688655 | 0.15 |
ENST00000461930.1
ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2
|
profilin 2 |
| chr1_-_236046872 | 0.15 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr5_-_76382989 | 0.14 |
ENST00000511587.1
|
ZBED3
|
zinc finger, BED-type containing 3 |
| chr9_+_74526384 | 0.14 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr16_+_53164833 | 0.13 |
ENST00000564845.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr5_+_74807886 | 0.13 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chr6_+_12958137 | 0.12 |
ENST00000457702.2
ENST00000379345.2 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr10_-_76868931 | 0.12 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr1_+_110036728 | 0.12 |
ENST00000369868.3
ENST00000430195.2 |
CYB561D1
|
cytochrome b561 family, member D1 |
| chr7_-_38403077 | 0.12 |
ENST00000426402.2
|
TRGV2
|
T cell receptor gamma variable 2 |
| chr19_-_6670128 | 0.11 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr11_-_14913765 | 0.10 |
ENST00000334636.5
|
CYP2R1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr4_-_17783135 | 0.10 |
ENST00000265018.3
|
FAM184B
|
family with sequence similarity 184, member B |
| chr5_-_115177247 | 0.10 |
ENST00000500945.2
|
ATG12
|
autophagy related 12 |
| chr12_+_27619743 | 0.09 |
ENST00000298876.4
ENST00000416383.1 |
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr5_+_112074029 | 0.09 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr15_+_64428529 | 0.09 |
ENST00000560861.1
|
SNX1
|
sorting nexin 1 |
| chr4_-_140216948 | 0.09 |
ENST00000265500.4
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr17_-_5487768 | 0.09 |
ENST00000269280.4
ENST00000345221.3 ENST00000262467.5 |
NLRP1
|
NLR family, pyrin domain containing 1 |
| chr18_-_33709268 | 0.09 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr11_-_18548426 | 0.09 |
ENST00000357193.3
ENST00000536719.1 |
TSG101
|
tumor susceptibility 101 |
| chrX_-_154563889 | 0.08 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr11_-_67980744 | 0.08 |
ENST00000401547.2
ENST00000453170.1 ENST00000304363.4 |
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr5_+_138609441 | 0.08 |
ENST00000509990.1
ENST00000506147.1 ENST00000512107.1 |
MATR3
|
matrin 3 |
| chr2_+_42396472 | 0.08 |
ENST00000318522.5
ENST00000402711.2 |
EML4
|
echinoderm microtubule associated protein like 4 |
| chrX_-_152343394 | 0.08 |
ENST00000370261.1
|
PNMA6B
|
paraneoplastic Ma antigen family member 6B (pseudogene) |
| chr2_-_40679148 | 0.08 |
ENST00000417271.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr5_+_74807581 | 0.08 |
ENST00000241436.4
ENST00000352007.5 |
POLK
|
polymerase (DNA directed) kappa |
| chr1_+_110036699 | 0.08 |
ENST00000496961.1
ENST00000533024.1 ENST00000310611.4 ENST00000527072.1 ENST00000420578.2 ENST00000528785.1 |
CYB561D1
|
cytochrome b561 family, member D1 |
| chr7_-_38394118 | 0.08 |
ENST00000390345.2
|
TRGV4
|
T cell receptor gamma variable 4 |
| chr14_-_92413353 | 0.07 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr14_-_90085458 | 0.07 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chr17_+_7531281 | 0.07 |
ENST00000575729.1
ENST00000340624.5 |
SHBG
|
sex hormone-binding globulin |
| chr19_-_39926268 | 0.07 |
ENST00000599705.1
|
RPS16
|
ribosomal protein S16 |
| chr2_-_25565377 | 0.07 |
ENST00000264709.3
ENST00000406659.3 |
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr1_+_159141397 | 0.07 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr6_+_123100853 | 0.07 |
ENST00000356535.4
|
FABP7
|
fatty acid binding protein 7, brain |
| chr19_-_2042065 | 0.06 |
ENST00000591588.1
ENST00000591142.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr2_-_154335300 | 0.06 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr6_+_30850697 | 0.06 |
ENST00000509639.1
ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_+_42396574 | 0.06 |
ENST00000401738.3
|
EML4
|
echinoderm microtubule associated protein like 4 |
| chr3_-_195163803 | 0.06 |
ENST00000326793.6
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr14_-_92413727 | 0.05 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr3_-_49131614 | 0.05 |
ENST00000450685.1
|
QRICH1
|
glutamine-rich 1 |
| chr16_-_68406161 | 0.05 |
ENST00000568373.1
ENST00000563226.1 |
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr5_+_76506706 | 0.05 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chrX_+_152224766 | 0.05 |
ENST00000370265.4
ENST00000447306.1 |
PNMA3
|
paraneoplastic Ma antigen 3 |
| chr5_+_75379224 | 0.05 |
ENST00000322285.7
|
SV2C
|
synaptic vesicle glycoprotein 2C |
| chr1_-_111148969 | 0.05 |
ENST00000316361.4
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr1_-_149908710 | 0.05 |
ENST00000439741.2
ENST00000361405.6 ENST00000406732.3 |
MTMR11
|
myotubularin related protein 11 |
| chr10_-_76868866 | 0.04 |
ENST00000607487.1
|
DUSP13
|
dual specificity phosphatase 13 |
| chr12_+_123949053 | 0.04 |
ENST00000350887.5
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr1_+_65210772 | 0.04 |
ENST00000371072.4
ENST00000294428.3 |
RAVER2
|
ribonucleoprotein, PTB-binding 2 |
| chr17_-_39623681 | 0.04 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr6_+_30850862 | 0.04 |
ENST00000504651.1
ENST00000512694.1 ENST00000515233.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr19_+_39926791 | 0.04 |
ENST00000594990.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr6_+_31939608 | 0.04 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr10_+_99332198 | 0.04 |
ENST00000307518.5
ENST00000298808.5 ENST00000370655.1 |
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chrX_+_11776278 | 0.03 |
ENST00000312196.4
ENST00000337339.2 |
MSL3
|
male-specific lethal 3 homolog (Drosophila) |
| chr5_-_74807418 | 0.03 |
ENST00000405807.4
ENST00000261415.7 |
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr17_+_61554829 | 0.03 |
ENST00000582627.1
|
ACE
|
angiotensin I converting enzyme |
| chr15_-_74659978 | 0.03 |
ENST00000541301.1
ENST00000416978.1 ENST00000268053.6 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr14_+_60715928 | 0.03 |
ENST00000395076.4
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr16_-_19896150 | 0.03 |
ENST00000570142.1
|
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr6_+_123100620 | 0.02 |
ENST00000368444.3
|
FABP7
|
fatty acid binding protein 7, brain |
| chr1_+_160160346 | 0.02 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr17_+_75542997 | 0.02 |
ENST00000592651.1
ENST00000590398.1 |
RP11-13K12.1
|
RP11-13K12.1 |
| chr20_+_54987168 | 0.02 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr5_+_170288856 | 0.02 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr9_+_72002837 | 0.02 |
ENST00000377216.3
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chrX_-_118827333 | 0.01 |
ENST00000360156.7
ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6
|
septin 6 |
| chr12_+_54718904 | 0.01 |
ENST00000262061.2
ENST00000549043.1 ENST00000552218.1 ENST00000553231.1 ENST00000552362.1 ENST00000455864.2 ENST00000416254.2 ENST00000549116.1 ENST00000551779.1 |
COPZ1
|
coatomer protein complex, subunit zeta 1 |
| chr9_+_140500087 | 0.01 |
ENST00000371421.4
|
ARRDC1
|
arrestin domain containing 1 |
| chr8_+_42196000 | 0.01 |
ENST00000518925.1
ENST00000538005.1 |
POLB
|
polymerase (DNA directed), beta |
| chr5_-_94417339 | 0.01 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr1_-_111148241 | 0.01 |
ENST00000440270.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr6_-_46138676 | 0.01 |
ENST00000371383.2
ENST00000230565.3 |
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative) |
| chr2_-_136594740 | 0.01 |
ENST00000264162.2
|
LCT
|
lactase |
| chr14_-_92414055 | 0.01 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr10_-_73533255 | 0.01 |
ENST00000394957.3
|
C10orf54
|
chromosome 10 open reading frame 54 |
| chr2_-_74570520 | 0.01 |
ENST00000394019.2
ENST00000346834.4 ENST00000359484.4 ENST00000423644.1 ENST00000377634.4 ENST00000436454.1 |
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chrX_-_152245978 | 0.01 |
ENST00000538162.2
|
PNMA6D
|
paraneoplastic Ma antigen family member 6D (pseudogene) |
| chr3_+_98250743 | 0.01 |
ENST00000284311.3
|
GPR15
|
G protein-coupled receptor 15 |
| chr19_+_52848659 | 0.01 |
ENST00000327920.8
|
ZNF610
|
zinc finger protein 610 |
| chr8_+_144718171 | 0.01 |
ENST00000526926.1
ENST00000458270.2 |
ZNF623
|
zinc finger protein 623 |
| chr1_+_155146318 | 0.01 |
ENST00000368385.4
ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46
|
tripartite motif containing 46 |
| chr6_+_44126545 | 0.00 |
ENST00000532171.1
ENST00000398776.1 ENST00000542245.1 |
CAPN11
|
calpain 11 |
| chr19_-_55549624 | 0.00 |
ENST00000417454.1
ENST00000310373.3 ENST00000333884.2 |
GP6
|
glycoprotein VI (platelet) |
| chr4_+_159236462 | 0.00 |
ENST00000460056.2
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr18_+_32073253 | 0.00 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr1_+_210001309 | 0.00 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr21_+_45209394 | 0.00 |
ENST00000497547.1
|
RRP1
|
ribosomal RNA processing 1 |
| chrX_+_152240819 | 0.00 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:2000397 | positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.1 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.0 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.3 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |