Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
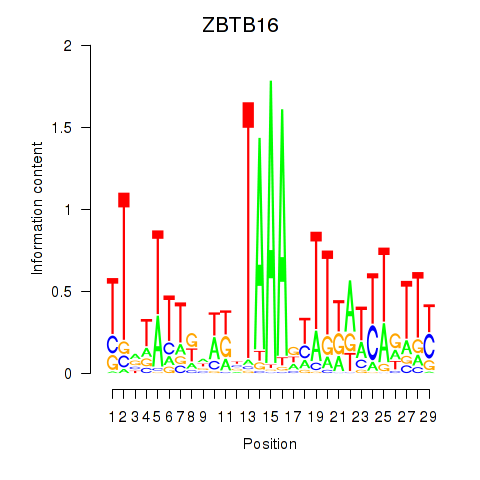
Results for ZBTB16
Z-value: 0.97
Transcription factors associated with ZBTB16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB16
|
ENSG00000109906.9 | zinc finger and BTB domain containing 16 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB16 | hg19_v2_chr11_+_113930291_113930339 | -0.99 | 1.5e-04 | Click! |
Activity profile of ZBTB16 motif
Sorted Z-values of ZBTB16 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_98347063 | 1.07 |
ENST00000443638.1
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr7_+_16828866 | 1.07 |
ENST00000597084.1
|
AC073333.1
|
Uncharacterized protein |
| chr1_+_33546714 | 0.92 |
ENST00000294517.6
ENST00000358680.3 ENST00000373443.3 ENST00000398167.1 |
ADC
|
arginine decarboxylase |
| chr13_-_101240985 | 0.84 |
ENST00000471912.1
|
GGACT
|
gamma-glutamylamine cyclotransferase |
| chr21_-_40817645 | 0.82 |
ENST00000438404.1
ENST00000358268.2 ENST00000411566.1 ENST00000451131.1 ENST00000418018.1 ENST00000415863.1 ENST00000426783.1 ENST00000288350.3 ENST00000485895.2 ENST00000448288.2 ENST00000456017.1 ENST00000434281.1 |
LCA5L
|
Leber congenital amaurosis 5-like |
| chr17_-_39341594 | 0.77 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr4_+_96012585 | 0.69 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr12_-_25150373 | 0.68 |
ENST00000549828.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr19_+_3762703 | 0.64 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr18_+_6256746 | 0.62 |
ENST00000578427.1
|
RP11-760N9.1
|
RP11-760N9.1 |
| chr12_-_10007448 | 0.62 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr19_+_42041860 | 0.62 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr1_+_43613566 | 0.61 |
ENST00000409396.1
|
FAM183A
|
family with sequence similarity 183, member A |
| chr7_-_32534850 | 0.55 |
ENST00000409952.3
ENST00000409909.3 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr10_+_62538089 | 0.51 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr12_+_97306295 | 0.51 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr9_-_15472730 | 0.49 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr5_+_180682720 | 0.48 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr19_+_20959098 | 0.47 |
ENST00000360204.5
ENST00000594534.1 |
ZNF66
|
zinc finger protein 66 |
| chr19_+_36239576 | 0.47 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr4_+_74301880 | 0.46 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr3_+_148415571 | 0.46 |
ENST00000497524.1
ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr2_-_188419078 | 0.45 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr4_+_26323764 | 0.45 |
ENST00000514730.1
ENST00000507574.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr4_+_95128748 | 0.45 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr14_+_67656110 | 0.44 |
ENST00000524532.1
ENST00000530728.1 |
FAM71D
|
family with sequence similarity 71, member D |
| chr16_-_70239683 | 0.44 |
ENST00000601706.1
|
AC009060.1
|
Uncharacterized protein |
| chr1_+_207277632 | 0.42 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr3_-_146262488 | 0.42 |
ENST00000487389.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr4_+_26324474 | 0.41 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr21_+_40817749 | 0.40 |
ENST00000380637.3
ENST00000380634.1 ENST00000458295.1 ENST00000440288.2 ENST00000380631.1 |
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr4_+_74606223 | 0.38 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr6_-_47445214 | 0.38 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr12_-_10601963 | 0.38 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr3_-_138312971 | 0.38 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
| chr1_-_228353112 | 0.37 |
ENST00000366713.1
|
IBA57-AS1
|
IBA57 antisense RNA 1 (head to head) |
| chr2_+_192543694 | 0.37 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr5_-_24645078 | 0.36 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr1_+_204494618 | 0.36 |
ENST00000367180.1
ENST00000391947.2 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr12_-_74686314 | 0.36 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr19_+_46531127 | 0.36 |
ENST00000601033.1
|
CTC-344H19.4
|
CTC-344H19.4 |
| chr17_+_19314432 | 0.35 |
ENST00000575165.2
|
RNF112
|
ring finger protein 112 |
| chr4_+_154622652 | 0.34 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr6_+_26273144 | 0.34 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr18_-_52626622 | 0.33 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr6_+_136172820 | 0.32 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr8_-_74495065 | 0.32 |
ENST00000523533.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr17_-_36997708 | 0.32 |
ENST00000398575.4
|
C17orf98
|
chromosome 17 open reading frame 98 |
| chr16_-_20817753 | 0.32 |
ENST00000389345.5
ENST00000300005.3 ENST00000357967.4 ENST00000569729.1 |
ERI2
|
ERI1 exoribonuclease family member 2 |
| chr7_+_7606497 | 0.32 |
ENST00000340080.4
ENST00000405785.1 ENST00000433635.1 |
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr3_-_93747425 | 0.32 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr15_-_54051831 | 0.32 |
ENST00000557913.1
ENST00000360509.5 |
WDR72
|
WD repeat domain 72 |
| chr20_-_33264886 | 0.31 |
ENST00000217446.3
ENST00000452740.2 ENST00000374820.2 |
PIGU
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr3_-_146262637 | 0.31 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr15_-_52944231 | 0.30 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr10_+_52094298 | 0.30 |
ENST00000595931.1
|
AC069547.1
|
HCG1745369; PRO3073; Uncharacterized protein |
| chr12_+_27849378 | 0.30 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr1_+_76251912 | 0.30 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr14_+_67831576 | 0.30 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr17_-_18266818 | 0.30 |
ENST00000583780.1
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr4_-_159094194 | 0.29 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr19_-_23869970 | 0.29 |
ENST00000601010.1
|
ZNF675
|
zinc finger protein 675 |
| chr12_+_58166726 | 0.28 |
ENST00000546504.1
|
RP11-571M6.15
|
Uncharacterized protein |
| chr3_-_138313161 | 0.28 |
ENST00000489254.1
ENST00000474781.1 ENST00000462419.1 ENST00000264982.3 |
CEP70
|
centrosomal protein 70kDa |
| chr20_-_34042558 | 0.27 |
ENST00000374372.1
|
GDF5
|
growth differentiation factor 5 |
| chr12_+_20968608 | 0.27 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr12_-_27090896 | 0.27 |
ENST00000539625.1
ENST00000538727.1 |
ASUN
|
asunder spermatogenesis regulator |
| chr14_+_58754751 | 0.27 |
ENST00000598233.1
|
AL132989.1
|
AL132989.1 |
| chr7_+_7606639 | 0.26 |
ENST00000433056.1
|
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr1_+_104159999 | 0.26 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr5_+_68860949 | 0.26 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr7_-_92777606 | 0.26 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr12_+_32832134 | 0.26 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr20_-_18477862 | 0.25 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr7_+_112120908 | 0.25 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr19_+_45690646 | 0.24 |
ENST00000591569.1
|
AC006126.3
|
Uncharacterized protein |
| chr10_+_104613980 | 0.23 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr12_+_133657461 | 0.23 |
ENST00000412146.2
ENST00000544426.1 ENST00000440984.2 ENST00000319849.3 ENST00000440550.2 |
ZNF140
|
zinc finger protein 140 |
| chr20_-_44298878 | 0.23 |
ENST00000324384.3
ENST00000356562.2 |
WFDC11
|
WAP four-disulfide core domain 11 |
| chr19_-_46145696 | 0.22 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr19_-_45004556 | 0.22 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr3_-_133969673 | 0.22 |
ENST00000427044.2
|
RYK
|
receptor-like tyrosine kinase |
| chr7_+_7606605 | 0.22 |
ENST00000456533.1
|
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr8_-_33370607 | 0.21 |
ENST00000360742.5
ENST00000523305.1 |
TTI2
|
TELO2 interacting protein 2 |
| chr8_+_66619277 | 0.21 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr22_+_29168652 | 0.21 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr2_+_42104692 | 0.21 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chrX_+_23925918 | 0.20 |
ENST00000379211.3
|
CXorf58
|
chromosome X open reading frame 58 |
| chr19_-_19932501 | 0.20 |
ENST00000540806.2
ENST00000590766.1 ENST00000587452.1 ENST00000545006.1 ENST00000590319.1 ENST00000587461.1 ENST00000450683.2 ENST00000443905.2 ENST00000590274.1 |
ZNF506
CTC-559E9.4
|
zinc finger protein 506 CTC-559E9.4 |
| chr12_-_53601055 | 0.20 |
ENST00000552972.1
ENST00000422257.3 ENST00000267082.5 |
ITGB7
|
integrin, beta 7 |
| chr3_-_169487617 | 0.20 |
ENST00000330368.2
|
ACTRT3
|
actin-related protein T3 |
| chr19_+_21265028 | 0.20 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr12_-_75905374 | 0.19 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr4_+_14113592 | 0.19 |
ENST00000502759.1
ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085
|
long intergenic non-protein coding RNA 1085 |
| chr19_-_55150343 | 0.19 |
ENST00000456337.1
|
AC009892.10
|
Uncharacterized protein |
| chr15_+_45028719 | 0.19 |
ENST00000560442.1
ENST00000558329.1 ENST00000561043.1 |
TRIM69
|
tripartite motif containing 69 |
| chr4_-_185275104 | 0.18 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr8_+_67687773 | 0.18 |
ENST00000518388.1
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr2_+_200472779 | 0.18 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr5_+_35856951 | 0.18 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr17_+_74463650 | 0.18 |
ENST00000392492.3
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr5_+_179135246 | 0.18 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr14_+_39703112 | 0.18 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr8_+_67039131 | 0.17 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr3_+_51663407 | 0.17 |
ENST00000432863.1
ENST00000296477.3 |
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr6_-_165723088 | 0.17 |
ENST00000230301.8
|
C6orf118
|
chromosome 6 open reading frame 118 |
| chr22_+_30821784 | 0.17 |
ENST00000407550.3
|
MTFP1
|
mitochondrial fission process 1 |
| chr12_-_68696652 | 0.16 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr15_-_54025300 | 0.16 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr17_-_46623441 | 0.16 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr1_+_76251879 | 0.15 |
ENST00000535300.1
ENST00000319942.3 |
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr4_-_39033963 | 0.15 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr3_-_93692781 | 0.15 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr5_-_54988559 | 0.15 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr2_+_105050794 | 0.15 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr4_+_106631966 | 0.15 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr3_+_184529948 | 0.15 |
ENST00000436792.2
ENST00000446204.2 ENST00000422105.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr4_-_88244049 | 0.15 |
ENST00000328546.4
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr14_-_45722360 | 0.14 |
ENST00000451174.1
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr3_+_184529929 | 0.14 |
ENST00000287546.4
ENST00000437079.3 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr19_+_42041537 | 0.14 |
ENST00000597199.1
|
AC006129.4
|
AC006129.4 |
| chrX_-_24045303 | 0.14 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr18_-_64271363 | 0.14 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr1_+_210501589 | 0.14 |
ENST00000413764.2
ENST00000541565.1 |
HHAT
|
hedgehog acyltransferase |
| chr17_-_40134339 | 0.14 |
ENST00000587727.1
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr19_-_53324884 | 0.14 |
ENST00000457749.2
ENST00000414252.2 ENST00000391783.2 |
ZNF28
|
zinc finger protein 28 |
| chr12_+_105380073 | 0.13 |
ENST00000552951.1
ENST00000280749.5 |
C12orf45
|
chromosome 12 open reading frame 45 |
| chr19_+_36142147 | 0.13 |
ENST00000590618.1
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr12_-_71533055 | 0.13 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr16_+_48657337 | 0.13 |
ENST00000568150.1
ENST00000564212.1 |
RP11-42I10.1
|
RP11-42I10.1 |
| chr19_+_42041702 | 0.13 |
ENST00000487420.2
|
AC006129.4
|
AC006129.4 |
| chr10_+_134258649 | 0.13 |
ENST00000392630.3
ENST00000321248.2 |
C10orf91
|
chromosome 10 open reading frame 91 |
| chr9_-_106087316 | 0.12 |
ENST00000411575.1
|
RP11-341A22.2
|
RP11-341A22.2 |
| chr22_-_38239808 | 0.12 |
ENST00000406423.1
ENST00000424350.1 ENST00000458278.2 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr19_+_21264943 | 0.12 |
ENST00000597424.1
|
ZNF714
|
zinc finger protein 714 |
| chr21_+_37692481 | 0.12 |
ENST00000400485.1
|
MORC3
|
MORC family CW-type zinc finger 3 |
| chr5_-_34919094 | 0.12 |
ENST00000341754.4
|
RAD1
|
RAD1 homolog (S. pombe) |
| chrX_-_2882296 | 0.12 |
ENST00000438544.1
ENST00000381134.3 ENST00000545496.1 |
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1) |
| chr10_+_97709725 | 0.12 |
ENST00000472454.2
|
RP11-248J23.6
|
Protein LOC100652732 |
| chr13_+_76123883 | 0.11 |
ENST00000377595.3
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr2_+_242289502 | 0.11 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr4_+_86699834 | 0.11 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr2_-_228244013 | 0.11 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr14_+_39703084 | 0.11 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr12_+_21284118 | 0.11 |
ENST00000256958.2
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1 |
| chr17_+_22022437 | 0.11 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr12_+_32832203 | 0.11 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr5_+_72509751 | 0.11 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr15_+_45028753 | 0.11 |
ENST00000338264.4
|
TRIM69
|
tripartite motif containing 69 |
| chr4_-_88244010 | 0.10 |
ENST00000302219.6
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr19_-_46195029 | 0.10 |
ENST00000588599.1
ENST00000585392.1 ENST00000590212.1 ENST00000587367.1 ENST00000391932.3 |
SNRPD2
|
small nuclear ribonucleoprotein D2 polypeptide 16.5kDa |
| chr12_+_50690489 | 0.10 |
ENST00000598429.1
|
AC140061.12
|
Uncharacterized protein |
| chr16_+_20817761 | 0.10 |
ENST00000568046.1
ENST00000261377.6 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr3_+_179065474 | 0.10 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr11_-_104972158 | 0.10 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr12_+_21207503 | 0.09 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chrX_+_118602363 | 0.09 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr5_-_176827631 | 0.09 |
ENST00000358571.2
|
PFN3
|
profilin 3 |
| chr2_+_172544011 | 0.09 |
ENST00000508530.1
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr3_-_156272924 | 0.08 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr3_+_160394940 | 0.08 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr4_-_99851766 | 0.08 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr15_-_50406680 | 0.08 |
ENST00000559829.1
|
ATP8B4
|
ATPase, class I, type 8B, member 4 |
| chr19_+_7011509 | 0.08 |
ENST00000377296.3
|
AC025278.1
|
Uncharacterized protein |
| chr20_+_31407692 | 0.07 |
ENST00000375571.5
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr1_-_44818599 | 0.07 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr7_-_16872932 | 0.07 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr14_+_52164820 | 0.07 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr9_+_131901661 | 0.07 |
ENST00000423100.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr1_+_100731749 | 0.07 |
ENST00000370128.4
ENST00000260563.4 |
RTCA
|
RNA 3'-terminal phosphate cyclase |
| chr19_-_56343353 | 0.07 |
ENST00000592953.1
ENST00000589093.1 |
NLRP11
|
NLR family, pyrin domain containing 11 |
| chr1_+_149239529 | 0.07 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr4_-_38784572 | 0.07 |
ENST00000308973.4
ENST00000361424.2 |
TLR10
|
toll-like receptor 10 |
| chr16_-_46649905 | 0.06 |
ENST00000569702.1
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr2_+_172543967 | 0.06 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr16_-_14109841 | 0.06 |
ENST00000576797.1
ENST00000575424.1 |
CTD-2135D7.5
|
CTD-2135D7.5 |
| chr1_+_170633047 | 0.06 |
ENST00000239461.6
ENST00000497230.2 |
PRRX1
|
paired related homeobox 1 |
| chr16_-_4524543 | 0.06 |
ENST00000573571.1
ENST00000404295.3 ENST00000574425.1 |
NMRAL1
|
NmrA-like family domain containing 1 |
| chr17_-_74351010 | 0.06 |
ENST00000435555.2
|
PRPSAP1
|
phosphoribosyl pyrophosphate synthetase-associated protein 1 |
| chr1_+_32608566 | 0.06 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr10_-_75226166 | 0.06 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr5_+_159848807 | 0.06 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr7_+_106505912 | 0.06 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr17_+_40925454 | 0.06 |
ENST00000253794.2
ENST00000590339.1 ENST00000589520.1 |
VPS25
|
vacuolar protein sorting 25 homolog (S. cerevisiae) |
| chr1_-_115292591 | 0.06 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr19_+_13135386 | 0.06 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr10_-_61495760 | 0.05 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr12_-_16761117 | 0.05 |
ENST00000538051.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr15_+_69365265 | 0.05 |
ENST00000415504.1
|
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr16_+_24549014 | 0.05 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr1_+_43613612 | 0.05 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr17_-_39183452 | 0.05 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr7_-_104909435 | 0.05 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr5_-_55412774 | 0.05 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr2_+_89901292 | 0.05 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr7_+_134464414 | 0.04 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr22_-_30953587 | 0.04 |
ENST00000453479.1
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr4_-_38784592 | 0.04 |
ENST00000502321.1
|
TLR10
|
toll-like receptor 10 |
| chr18_+_616711 | 0.04 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr8_+_104831554 | 0.04 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB16
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903423 | synaptic vesicle recycling via endosome(GO:0036466) positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.5 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.9 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.6 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.3 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.5 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.1 | 0.7 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 0.5 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.2 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.2 | GO:1904482 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.1 | 0.2 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.4 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.1 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.0 | 0.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 1.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.8 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.3 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.2 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.2 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.6 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.5 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.3 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) glial limiting end-foot(GO:0097451) |
| 0.1 | 0.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.9 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 0.5 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.5 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.2 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |