Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
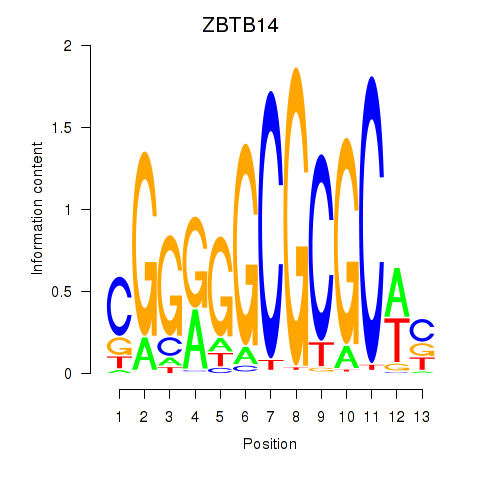
Results for ZBTB14
Z-value: 0.75
Transcription factors associated with ZBTB14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB14
|
ENSG00000198081.6 | zinc finger and BTB domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB14 | hg19_v2_chr18_-_5295679_5295734 | -0.38 | 4.5e-01 | Click! |
Activity profile of ZBTB14 motif
Sorted Z-values of ZBTB14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_49463620 | 0.60 |
ENST00000550675.1
|
RHEBL1
|
Ras homolog enriched in brain like 1 |
| chr8_-_42752418 | 0.49 |
ENST00000524954.1
|
RNF170
|
ring finger protein 170 |
| chr3_-_185826718 | 0.49 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr3_-_118753792 | 0.44 |
ENST00000480431.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr19_+_45596398 | 0.40 |
ENST00000544069.2
|
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr22_+_46972975 | 0.39 |
ENST00000431155.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr19_+_35634146 | 0.39 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr2_+_74212073 | 0.38 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25 |
| chr5_+_98264867 | 0.36 |
ENST00000513175.1
|
CTD-2007H13.3
|
CTD-2007H13.3 |
| chr2_+_18059906 | 0.34 |
ENST00000304101.4
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr3_-_185826286 | 0.34 |
ENST00000537818.1
ENST00000422039.1 ENST00000434744.1 |
ETV5
|
ets variant 5 |
| chr8_+_61592073 | 0.33 |
ENST00000526846.1
|
CHD7
|
chromodomain helicase DNA binding protein 7 |
| chr14_-_23770683 | 0.32 |
ENST00000561437.1
ENST00000559942.1 ENST00000560913.1 ENST00000559314.1 ENST00000558058.1 |
PPP1R3E
|
protein phosphatase 1, regulatory subunit 3E |
| chr11_+_65122216 | 0.32 |
ENST00000309880.5
|
TIGD3
|
tigger transposable element derived 3 |
| chr20_+_9049742 | 0.32 |
ENST00000437503.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr20_-_61847455 | 0.30 |
ENST00000370334.4
|
YTHDF1
|
YTH domain family, member 1 |
| chr19_-_36247910 | 0.29 |
ENST00000587965.1
ENST00000004982.3 |
HSPB6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr22_-_31741757 | 0.29 |
ENST00000215919.3
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr3_+_14989076 | 0.28 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr11_+_113930955 | 0.27 |
ENST00000535700.1
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr19_-_40732594 | 0.27 |
ENST00000430325.2
ENST00000433940.1 |
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr1_-_33336414 | 0.26 |
ENST00000373471.3
ENST00000609187.1 |
FNDC5
|
fibronectin type III domain containing 5 |
| chr3_-_9595480 | 0.26 |
ENST00000287585.6
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4 |
| chr20_-_56284816 | 0.26 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr17_-_67323385 | 0.26 |
ENST00000588665.1
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr11_+_57435441 | 0.25 |
ENST00000528177.1
|
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr19_+_44037334 | 0.25 |
ENST00000314228.5
|
ZNF575
|
zinc finger protein 575 |
| chr3_-_141868293 | 0.25 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr11_-_119234876 | 0.25 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr16_+_67197288 | 0.25 |
ENST00000264009.8
ENST00000421453.1 |
HSF4
|
heat shock transcription factor 4 |
| chr12_-_51663682 | 0.24 |
ENST00000603838.1
|
SMAGP
|
small cell adhesion glycoprotein |
| chr12_-_90103077 | 0.24 |
ENST00000551310.1
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr17_+_59477233 | 0.24 |
ENST00000240328.3
|
TBX2
|
T-box 2 |
| chrX_+_70364667 | 0.23 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr19_+_18942720 | 0.23 |
ENST00000262803.5
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast) |
| chr11_-_30607819 | 0.23 |
ENST00000448418.2
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr17_-_42277203 | 0.23 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr12_-_125052010 | 0.23 |
ENST00000458234.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr14_+_91526668 | 0.22 |
ENST00000521334.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr1_-_223537401 | 0.22 |
ENST00000343846.3
ENST00000454695.2 ENST00000484758.2 |
SUSD4
|
sushi domain containing 4 |
| chr11_-_72852320 | 0.22 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr3_+_61547585 | 0.22 |
ENST00000295874.10
ENST00000474889.1 |
PTPRG
|
protein tyrosine phosphatase, receptor type, G |
| chr22_+_24129138 | 0.21 |
ENST00000417137.1
ENST00000344921.6 ENST00000263121.7 ENST00000407422.3 ENST00000407082.3 |
SMARCB1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 |
| chr12_-_26278030 | 0.21 |
ENST00000242728.4
|
BHLHE41
|
basic helix-loop-helix family, member e41 |
| chr19_+_6464502 | 0.21 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr5_+_176560595 | 0.21 |
ENST00000508896.1
|
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr19_-_6199521 | 0.21 |
ENST00000592883.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr19_+_35521616 | 0.21 |
ENST00000595652.1
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr12_-_99288536 | 0.21 |
ENST00000549797.1
ENST00000333732.7 ENST00000341752.7 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr7_-_139876812 | 0.20 |
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr19_+_18942761 | 0.20 |
ENST00000599848.1
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast) |
| chr5_+_133859996 | 0.20 |
ENST00000512386.1
|
PHF15
|
jade family PHD finger 2 |
| chr11_-_65625014 | 0.20 |
ENST00000534784.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr9_+_103189405 | 0.20 |
ENST00000395067.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr2_+_204192942 | 0.20 |
ENST00000295851.5
ENST00000261017.5 |
ABI2
|
abl-interactor 2 |
| chr18_-_74534232 | 0.20 |
ENST00000585258.1
|
RP11-162A12.2
|
Uncharacterized protein |
| chr6_-_13487825 | 0.19 |
ENST00000603223.1
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr9_-_102582155 | 0.19 |
ENST00000427039.1
|
RP11-554F20.1
|
RP11-554F20.1 |
| chr9_+_6716478 | 0.19 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3 |
| chr21_-_46237883 | 0.19 |
ENST00000397893.3
|
SUMO3
|
small ubiquitin-like modifier 3 |
| chr9_+_132099158 | 0.19 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr4_-_140477353 | 0.19 |
ENST00000406354.1
ENST00000506866.2 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr16_+_30710462 | 0.19 |
ENST00000262518.4
ENST00000395059.2 ENST00000344771.4 |
SRCAP
|
Snf2-related CREBBP activator protein |
| chr10_-_735553 | 0.18 |
ENST00000280886.6
ENST00000423550.1 |
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr1_+_228674762 | 0.18 |
ENST00000305943.7
|
RNF187
|
ring finger protein 187 |
| chr6_+_111408698 | 0.18 |
ENST00000368851.5
|
SLC16A10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr16_+_84402098 | 0.18 |
ENST00000262429.4
ENST00000416219.2 |
ATP2C2
|
ATPase, Ca++ transporting, type 2C, member 2 |
| chr22_+_29999647 | 0.18 |
ENST00000334961.7
ENST00000353887.4 |
NF2
|
neurofibromin 2 (merlin) |
| chr1_+_36038971 | 0.18 |
ENST00000373235.3
|
TFAP2E
|
transcription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) |
| chr19_+_1407733 | 0.18 |
ENST00000592453.1
|
DAZAP1
|
DAZ associated protein 1 |
| chr20_-_62462566 | 0.18 |
ENST00000245663.4
ENST00000302995.2 |
ZBTB46
|
zinc finger and BTB domain containing 46 |
| chr14_+_89029323 | 0.18 |
ENST00000554602.1
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr15_+_91643442 | 0.18 |
ENST00000394232.1
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr14_-_104313824 | 0.17 |
ENST00000553739.1
ENST00000202556.9 |
PPP1R13B
|
protein phosphatase 1, regulatory subunit 13B |
| chr1_-_21948906 | 0.17 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr9_-_127703333 | 0.17 |
ENST00000373555.4
|
GOLGA1
|
golgin A1 |
| chr16_-_79633799 | 0.17 |
ENST00000569649.1
|
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr16_+_31085714 | 0.17 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr20_-_62610982 | 0.17 |
ENST00000369886.3
ENST00000450107.1 |
SAMD10
|
sterile alpha motif domain containing 10 |
| chr9_+_101705893 | 0.17 |
ENST00000375001.3
|
COL15A1
|
collagen, type XV, alpha 1 |
| chr5_+_172068232 | 0.17 |
ENST00000520919.1
ENST00000522853.1 ENST00000369800.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr14_+_60716276 | 0.17 |
ENST00000528241.2
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr2_-_220094031 | 0.17 |
ENST00000443140.1
ENST00000432520.1 ENST00000409618.1 |
ATG9A
|
autophagy related 9A |
| chr1_+_41249539 | 0.17 |
ENST00000347132.5
ENST00000509682.2 |
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4 |
| chr8_+_55370487 | 0.17 |
ENST00000297316.4
|
SOX17
|
SRY (sex determining region Y)-box 17 |
| chr7_-_44365216 | 0.17 |
ENST00000358707.3
ENST00000457475.1 ENST00000440254.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr3_+_170136642 | 0.17 |
ENST00000064724.3
ENST00000486975.1 |
CLDN11
|
claudin 11 |
| chr8_+_99076750 | 0.17 |
ENST00000545282.1
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr16_+_25703274 | 0.17 |
ENST00000331351.5
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr21_+_44394742 | 0.16 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr19_+_18451439 | 0.16 |
ENST00000597431.2
|
PGPEP1
|
pyroglutamyl-peptidase I |
| chr17_+_43972010 | 0.16 |
ENST00000334239.8
ENST00000446361.3 |
MAPT
|
microtubule-associated protein tau |
| chr11_-_74660065 | 0.16 |
ENST00000525407.1
ENST00000528219.1 ENST00000531852.1 |
XRRA1
|
X-ray radiation resistance associated 1 |
| chr11_+_46368956 | 0.16 |
ENST00000543978.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr7_-_72972319 | 0.16 |
ENST00000223368.2
|
BCL7B
|
B-cell CLL/lymphoma 7B |
| chr22_+_29999545 | 0.16 |
ENST00000413209.2
ENST00000347330.5 ENST00000338641.4 ENST00000403435.1 ENST00000361452.4 ENST00000403999.3 |
NF2
|
neurofibromin 2 (merlin) |
| chr19_+_6464243 | 0.16 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr1_+_6845578 | 0.16 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr17_+_4736812 | 0.15 |
ENST00000453408.3
|
MINK1
|
misshapen-like kinase 1 |
| chr6_-_90121789 | 0.15 |
ENST00000359203.3
|
RRAGD
|
Ras-related GTP binding D |
| chr20_+_45523227 | 0.15 |
ENST00000327619.5
ENST00000357410.3 |
EYA2
|
eyes absent homolog 2 (Drosophila) |
| chr12_-_124457371 | 0.15 |
ENST00000238156.3
ENST00000545037.1 |
CCDC92
|
coiled-coil domain containing 92 |
| chr13_+_49794474 | 0.15 |
ENST00000218721.1
ENST00000398307.1 |
MLNR
|
motilin receptor |
| chr4_-_1242463 | 0.15 |
ENST00000513420.1
|
CTBP1
|
C-terminal binding protein 1 |
| chr15_-_67813924 | 0.15 |
ENST00000559298.1
|
IQCH-AS1
|
IQCH antisense RNA 1 |
| chr4_-_24914508 | 0.15 |
ENST00000504487.1
|
CCDC149
|
coiled-coil domain containing 149 |
| chr7_-_27135591 | 0.15 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr12_+_1100370 | 0.15 |
ENST00000543086.3
ENST00000546231.2 ENST00000397203.2 |
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr1_+_44444865 | 0.15 |
ENST00000372324.1
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr17_-_38520067 | 0.15 |
ENST00000337376.4
ENST00000578689.1 |
GJD3
|
gap junction protein, delta 3, 31.9kDa |
| chr19_-_11591848 | 0.15 |
ENST00000359227.3
|
ELAVL3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr5_+_142149932 | 0.15 |
ENST00000274498.4
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr2_+_204193101 | 0.15 |
ENST00000430418.1
ENST00000424558.1 ENST00000261016.6 |
ABI2
|
abl-interactor 2 |
| chr10_-_105615164 | 0.15 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr14_+_24521202 | 0.15 |
ENST00000334420.7
ENST00000342740.5 |
LRRC16B
|
leucine rich repeat containing 16B |
| chr5_-_178054014 | 0.15 |
ENST00000520957.1
|
CLK4
|
CDC-like kinase 4 |
| chr11_-_134095335 | 0.15 |
ENST00000534227.1
ENST00000532445.1 |
NCAPD3
|
non-SMC condensin II complex, subunit D3 |
| chr20_+_306221 | 0.15 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr20_+_32077880 | 0.14 |
ENST00000342704.6
ENST00000375279.2 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr6_-_17987694 | 0.14 |
ENST00000378814.5
ENST00000378843.2 ENST00000378826.2 ENST00000378816.5 ENST00000259711.6 ENST00000502704.1 |
KIF13A
|
kinesin family member 13A |
| chr9_-_140196703 | 0.14 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chr20_+_306177 | 0.14 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr19_+_56111680 | 0.14 |
ENST00000301073.3
|
ZNF524
|
zinc finger protein 524 |
| chr16_-_90038866 | 0.14 |
ENST00000314994.3
|
CENPBD1
|
CENPB DNA-binding domains containing 1 |
| chr19_+_1407517 | 0.14 |
ENST00000336761.6
ENST00000233078.4 |
DAZAP1
|
DAZ associated protein 1 |
| chr2_+_64681103 | 0.14 |
ENST00000464281.1
|
LGALSL
|
lectin, galactoside-binding-like |
| chr19_+_1941117 | 0.14 |
ENST00000255641.8
|
CSNK1G2
|
casein kinase 1, gamma 2 |
| chr14_+_105781102 | 0.14 |
ENST00000547217.1
|
PACS2
|
phosphofurin acidic cluster sorting protein 2 |
| chr16_+_71660052 | 0.14 |
ENST00000567566.1
|
MARVELD3
|
MARVEL domain containing 3 |
| chr9_+_133539981 | 0.14 |
ENST00000253008.2
|
PRDM12
|
PR domain containing 12 |
| chr11_-_61348292 | 0.14 |
ENST00000539008.1
ENST00000540677.1 ENST00000542836.1 ENST00000542670.1 ENST00000535826.1 ENST00000545053.1 |
SYT7
|
synaptotagmin VII |
| chr17_-_27229948 | 0.14 |
ENST00000426464.2
|
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr1_+_16174280 | 0.14 |
ENST00000375759.3
|
SPEN
|
spen family transcriptional repressor |
| chr13_+_100634004 | 0.14 |
ENST00000376335.3
|
ZIC2
|
Zic family member 2 |
| chr9_+_103189536 | 0.14 |
ENST00000374885.1
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr17_-_42907564 | 0.13 |
ENST00000592524.1
|
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr19_+_55987998 | 0.13 |
ENST00000591164.1
|
ZNF628
|
zinc finger protein 628 |
| chr17_-_42908155 | 0.13 |
ENST00000426548.1
ENST00000590758.1 ENST00000591424.1 |
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr19_-_55628700 | 0.13 |
ENST00000592993.1
|
PPP1R12C
|
protein phosphatase 1, regulatory subunit 12C |
| chr1_+_46269248 | 0.13 |
ENST00000361297.2
ENST00000372009.2 |
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr1_+_111889212 | 0.13 |
ENST00000369737.4
|
PIFO
|
primary cilia formation |
| chr2_-_95831158 | 0.13 |
ENST00000447814.1
|
ZNF514
|
zinc finger protein 514 |
| chr1_-_29508499 | 0.13 |
ENST00000373795.4
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr4_-_146859787 | 0.13 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr3_+_14989186 | 0.13 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr6_+_157099036 | 0.13 |
ENST00000350026.5
ENST00000346085.5 ENST00000367148.1 ENST00000275248.4 |
ARID1B
|
AT rich interactive domain 1B (SWI1-like) |
| chr19_+_5904866 | 0.13 |
ENST00000339485.3
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr14_+_64319669 | 0.13 |
ENST00000358025.3
ENST00000357395.3 ENST00000344113.4 ENST00000341472.5 ENST00000356081.3 |
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr12_+_108956355 | 0.13 |
ENST00000535729.1
ENST00000431221.2 ENST00000547005.1 ENST00000311893.9 ENST00000392807.4 ENST00000338291.4 ENST00000539593.1 |
ISCU
|
iron-sulfur cluster assembly enzyme |
| chr5_-_524445 | 0.13 |
ENST00000514375.1
ENST00000264938.3 |
SLC9A3
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 |
| chr2_+_220094479 | 0.13 |
ENST00000323348.5
ENST00000453432.1 ENST00000409849.1 ENST00000416565.1 ENST00000410034.3 ENST00000447157.1 |
ANKZF1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr6_+_31126291 | 0.13 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr2_-_128145498 | 0.13 |
ENST00000409179.2
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr2_+_181845532 | 0.13 |
ENST00000602475.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr2_+_74741569 | 0.12 |
ENST00000233638.7
|
TLX2
|
T-cell leukemia homeobox 2 |
| chr16_+_5083950 | 0.12 |
ENST00000588623.1
|
ALG1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr16_+_30709530 | 0.12 |
ENST00000411466.2
|
SRCAP
|
Snf2-related CREBBP activator protein |
| chr11_-_45687128 | 0.12 |
ENST00000308064.2
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr22_+_50609150 | 0.12 |
ENST00000159647.5
ENST00000395842.2 |
PANX2
|
pannexin 2 |
| chr14_+_75894714 | 0.12 |
ENST00000559060.1
|
JDP2
|
Jun dimerization protein 2 |
| chrX_+_48554986 | 0.12 |
ENST00000376687.3
ENST00000453214.2 |
SUV39H1
|
suppressor of variegation 3-9 homolog 1 (Drosophila) |
| chr22_-_22090064 | 0.12 |
ENST00000339468.3
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chr1_-_2323140 | 0.12 |
ENST00000378531.3
ENST00000378529.3 |
MORN1
|
MORN repeat containing 1 |
| chr6_-_38607673 | 0.12 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr17_+_12693001 | 0.12 |
ENST00000262444.9
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr9_-_35072585 | 0.12 |
ENST00000448530.1
|
VCP
|
valosin containing protein |
| chrX_-_3631635 | 0.12 |
ENST00000262848.5
|
PRKX
|
protein kinase, X-linked |
| chr14_+_74003818 | 0.12 |
ENST00000311148.4
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr9_+_139971921 | 0.12 |
ENST00000409858.3
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 |
| chr12_-_49463753 | 0.12 |
ENST00000301068.6
|
RHEBL1
|
Ras homolog enriched in brain like 1 |
| chr3_+_49449636 | 0.12 |
ENST00000273590.3
|
TCTA
|
T-cell leukemia translocation altered |
| chr16_+_66914264 | 0.12 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr5_+_133861790 | 0.12 |
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2 |
| chr17_+_40811283 | 0.12 |
ENST00000251412.7
|
TUBG2
|
tubulin, gamma 2 |
| chr22_+_39795746 | 0.12 |
ENST00000216160.6
ENST00000331454.3 |
TAB1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr12_+_1100449 | 0.12 |
ENST00000360905.4
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr15_-_78111788 | 0.12 |
ENST00000566711.1
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr20_+_57466629 | 0.12 |
ENST00000371081.1
ENST00000338783.6 |
GNAS
|
GNAS complex locus |
| chr17_-_40761375 | 0.12 |
ENST00000543197.1
ENST00000309428.5 |
FAM134C
|
family with sequence similarity 134, member C |
| chr12_+_133067157 | 0.12 |
ENST00000261673.6
|
FBRSL1
|
fibrosin-like 1 |
| chr12_-_124457257 | 0.12 |
ENST00000545891.1
|
CCDC92
|
coiled-coil domain containing 92 |
| chr19_-_44809121 | 0.12 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr15_-_43882353 | 0.12 |
ENST00000453080.1
ENST00000360301.4 ENST00000360135.4 ENST00000417085.1 ENST00000431962.1 ENST00000334933.4 ENST00000381879.4 ENST00000420765.1 |
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr2_-_230786378 | 0.12 |
ENST00000430954.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr18_+_74534479 | 0.12 |
ENST00000320610.9
|
ZNF236
|
zinc finger protein 236 |
| chr2_+_204193129 | 0.12 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chr6_+_41606176 | 0.12 |
ENST00000441667.1
ENST00000230321.6 ENST00000373050.4 ENST00000446650.1 ENST00000435476.1 |
MDFI
|
MyoD family inhibitor |
| chr12_-_31479107 | 0.12 |
ENST00000542983.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr19_+_45596218 | 0.12 |
ENST00000421905.1
ENST00000221462.4 |
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr9_+_6758109 | 0.12 |
ENST00000536108.1
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr16_-_75285762 | 0.12 |
ENST00000564028.1
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr1_-_151319654 | 0.12 |
ENST00000430227.1
ENST00000412774.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr16_+_90086002 | 0.11 |
ENST00000563936.1
ENST00000536122.1 ENST00000561675.1 |
GAS8
|
growth arrest-specific 8 |
| chr21_+_38338737 | 0.11 |
ENST00000430068.1
|
AP000704.5
|
AP000704.5 |
| chr22_-_20104700 | 0.11 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr9_+_103189660 | 0.11 |
ENST00000374886.3
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr6_-_39082922 | 0.11 |
ENST00000229903.4
|
SAYSD1
|
SAYSVFN motif domain containing 1 |
| chr7_+_29846099 | 0.11 |
ENST00000409123.1
|
WIPF3
|
WAS/WASL interacting protein family, member 3 |
| chr5_+_142149955 | 0.11 |
ENST00000378004.3
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr3_+_52279902 | 0.11 |
ENST00000457454.1
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB14
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.4 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.1 | 0.3 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 | 0.2 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.2 | GO:0072387 | flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.1 | 0.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.2 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 0.2 | GO:0042662 | negative regulation of mesodermal cell fate specification(GO:0042662) regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) gall bladder development(GO:0061010) |
| 0.1 | 0.1 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.2 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.1 | 0.2 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.3 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.2 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.2 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.3 | GO:0051510 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.3 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.3 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.1 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.1 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 0.1 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.6 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.3 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.3 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.2 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.1 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.1 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.1 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.4 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.0 | GO:0071594 | T cell differentiation in thymus(GO:0033077) regulation of T cell differentiation in thymus(GO:0033081) thymocyte aggregation(GO:0071594) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.1 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.0 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.2 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.2 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.0 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.2 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.0 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.2 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.1 | GO:0071333 | cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.0 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.2 | GO:0010452 | regulation of histone H3-K36 methylation(GO:0000414) histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.1 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.0 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.0 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.0 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.1 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.0 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.0 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.0 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.1 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.0 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.0 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.2 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.0 | GO:1904806 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.1 | GO:0005940 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0043260 | laminin-1 complex(GO:0005606) laminin-11 complex(GO:0043260) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.0 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.1 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0042356 | GDP-4-dehydro-D-rhamnose reductase activity(GO:0042356) GDP-L-fucose synthase activity(GO:0050577) |
| 0.0 | 0.2 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.0 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.3 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.1 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.0 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.0 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |