Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
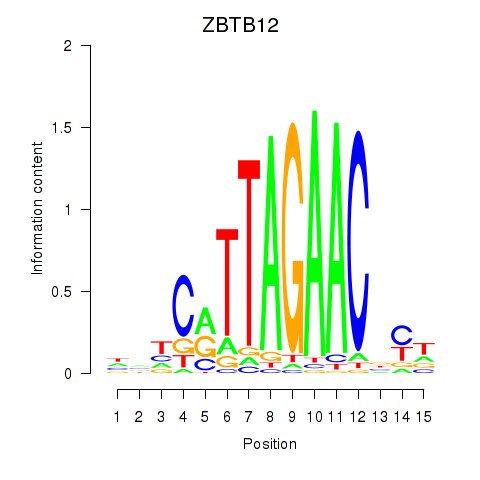
Results for ZBTB12
Z-value: 0.31
Transcription factors associated with ZBTB12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB12
|
ENSG00000204366.3 | zinc finger and BTB domain containing 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB12 | hg19_v2_chr6_-_31869769_31869880 | 0.22 | 6.7e-01 | Click! |
Activity profile of ZBTB12 motif
Sorted Z-values of ZBTB12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_54215029 | 0.32 |
ENST00000435813.2
|
RP11-346D6.6
|
RP11-346D6.6 |
| chr14_-_62162541 | 0.30 |
ENST00000557544.1
|
HIF1A-AS1
|
HIF1A antisense RNA 1 |
| chr12_-_123459105 | 0.26 |
ENST00000543935.1
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr14_+_32476072 | 0.20 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr14_+_20187174 | 0.19 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr1_+_158979680 | 0.19 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_158979792 | 0.18 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_158979686 | 0.18 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr12_-_46766577 | 0.17 |
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2 |
| chr10_+_63422695 | 0.16 |
ENST00000330194.2
ENST00000389639.3 |
C10orf107
|
chromosome 10 open reading frame 107 |
| chr12_+_95612006 | 0.15 |
ENST00000551311.1
ENST00000546445.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr11_-_10920838 | 0.15 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr15_-_35280426 | 0.15 |
ENST00000559564.1
ENST00000356321.4 |
ZNF770
|
zinc finger protein 770 |
| chr1_+_155583012 | 0.14 |
ENST00000462250.2
|
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chrX_-_122866874 | 0.14 |
ENST00000245838.8
ENST00000355725.4 |
THOC2
|
THO complex 2 |
| chr13_+_97928395 | 0.13 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr8_-_102987548 | 0.13 |
ENST00000520690.1
|
NCALD
|
neurocalcin delta |
| chr1_-_94344754 | 0.13 |
ENST00000436063.2
|
DNTTIP2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr19_-_36019123 | 0.13 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr14_-_50154921 | 0.13 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr1_+_61869748 | 0.13 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr8_+_38758737 | 0.12 |
ENST00000521746.1
ENST00000420274.1 |
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr15_+_40453204 | 0.12 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr6_-_114292449 | 0.12 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr10_+_76969909 | 0.11 |
ENST00000298468.5
ENST00000543351.1 |
VDAC2
|
voltage-dependent anion channel 2 |
| chr14_-_54908043 | 0.11 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr18_-_32870148 | 0.11 |
ENST00000589178.1
ENST00000333206.5 ENST00000592278.1 ENST00000592211.1 ENST00000420878.3 ENST00000383091.2 ENST00000586922.2 |
ZSCAN30
RP11-158H5.7
|
zinc finger and SCAN domain containing 30 RP11-158H5.7 |
| chrX_+_133930798 | 0.10 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr3_-_149051194 | 0.10 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr13_+_45694583 | 0.10 |
ENST00000340473.6
|
GTF2F2
|
general transcription factor IIF, polypeptide 2, 30kDa |
| chr4_-_99851766 | 0.09 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr15_+_66797627 | 0.09 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr15_+_66797455 | 0.09 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr16_+_31724552 | 0.09 |
ENST00000539915.1
ENST00000316491.9 ENST00000399681.3 ENST00000398696.3 ENST00000534369.1 |
ZNF720
|
zinc finger protein 720 |
| chr7_-_102985035 | 0.09 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr15_-_55562451 | 0.09 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr11_+_46332679 | 0.08 |
ENST00000530518.1
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr3_-_149051444 | 0.08 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr14_-_70826438 | 0.08 |
ENST00000389912.6
|
COX16
|
COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) |
| chr5_-_96518907 | 0.08 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr4_+_26859300 | 0.08 |
ENST00000494628.2
|
STIM2
|
stromal interaction molecule 2 |
| chrX_-_133931164 | 0.07 |
ENST00000370790.1
ENST00000298090.6 |
FAM122B
|
family with sequence similarity 122B |
| chrX_+_133371077 | 0.07 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr10_-_12084770 | 0.07 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr7_+_23637763 | 0.07 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr9_-_26946981 | 0.07 |
ENST00000523212.1
|
PLAA
|
phospholipase A2-activating protein |
| chr2_+_190722119 | 0.07 |
ENST00000452382.1
|
PMS1
|
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| chr9_+_127615733 | 0.07 |
ENST00000373574.1
|
WDR38
|
WD repeat domain 38 |
| chr8_-_33330595 | 0.07 |
ENST00000524021.1
ENST00000335589.3 |
FUT10
|
fucosyltransferase 10 (alpha (1,3) fucosyltransferase) |
| chrX_-_102942961 | 0.07 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr17_-_46178527 | 0.06 |
ENST00000393408.3
|
CBX1
|
chromobox homolog 1 |
| chr12_+_14572070 | 0.06 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr8_-_30670384 | 0.06 |
ENST00000221138.4
ENST00000518243.1 |
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr12_-_13248598 | 0.06 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr11_+_74303612 | 0.06 |
ENST00000527458.1
ENST00000532497.1 ENST00000530511.1 |
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr11_+_74303575 | 0.06 |
ENST00000263681.2
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr10_+_5454505 | 0.06 |
ENST00000355029.4
|
NET1
|
neuroepithelial cell transforming 1 |
| chr5_-_148929848 | 0.05 |
ENST00000504676.1
ENST00000515435.1 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr5_+_68513557 | 0.05 |
ENST00000256441.4
|
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr1_-_152779104 | 0.05 |
ENST00000606576.1
ENST00000368768.1 |
LCE1C
|
late cornified envelope 1C |
| chr1_+_35220613 | 0.05 |
ENST00000338513.1
|
GJB5
|
gap junction protein, beta 5, 31.1kDa |
| chr11_-_2170786 | 0.05 |
ENST00000300632.5
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr12_-_133532864 | 0.05 |
ENST00000536932.1
ENST00000360187.4 ENST00000392321.3 |
CHFR
ZNF605
|
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase zinc finger protein 605 |
| chr19_+_58258164 | 0.05 |
ENST00000317178.5
|
ZNF776
|
zinc finger protein 776 |
| chr15_-_49912987 | 0.05 |
ENST00000560246.1
ENST00000558594.1 |
FAM227B
|
family with sequence similarity 227, member B |
| chr18_-_57364588 | 0.05 |
ENST00000439986.4
|
CCBE1
|
collagen and calcium binding EGF domains 1 |
| chr7_-_93520259 | 0.05 |
ENST00000222543.5
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr10_+_180405 | 0.05 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr1_+_100598691 | 0.04 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr13_-_37633743 | 0.04 |
ENST00000497318.1
ENST00000475892.1 ENST00000356185.3 ENST00000350612.6 ENST00000542180.1 ENST00000360252.4 |
SUPT20H
|
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr2_+_234545092 | 0.04 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr17_-_5487768 | 0.04 |
ENST00000269280.4
ENST00000345221.3 ENST00000262467.5 |
NLRP1
|
NLR family, pyrin domain containing 1 |
| chr11_+_63449045 | 0.04 |
ENST00000354497.4
|
RTN3
|
reticulon 3 |
| chr14_+_96949319 | 0.04 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr16_-_30134441 | 0.04 |
ENST00000395200.1
|
MAPK3
|
mitogen-activated protein kinase 3 |
| chr1_+_11838988 | 0.04 |
ENST00000444493.1
|
C1orf167
|
chromosome 1 open reading frame 167 |
| chr16_-_30134524 | 0.04 |
ENST00000395202.1
ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr12_+_54378923 | 0.04 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr5_+_148786423 | 0.04 |
ENST00000505254.2
ENST00000602964.1 ENST00000519898.1 |
MIR143HG
|
MIR143 host gene (non-protein coding) |
| chr7_-_27169801 | 0.04 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr7_+_12544025 | 0.04 |
ENST00000443874.1
ENST00000424453.1 |
AC005281.1
|
AC005281.1 |
| chr2_+_197577841 | 0.04 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr1_+_46049706 | 0.04 |
ENST00000527470.1
ENST00000525515.1 ENST00000537798.1 ENST00000402363.3 ENST00000528238.1 ENST00000350030.3 ENST00000470768.1 ENST00000372052.4 ENST00000351223.3 |
NASP
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr10_+_57358750 | 0.04 |
ENST00000512524.2
|
MTRNR2L5
|
MT-RNR2-like 5 |
| chr4_-_87515202 | 0.04 |
ENST00000502302.1
ENST00000513186.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr1_-_109203997 | 0.04 |
ENST00000370032.5
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr11_+_64323156 | 0.04 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr11_+_64323098 | 0.03 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr4_+_9174899 | 0.03 |
ENST00000432515.1
|
FAM90A26
|
family with sequence similarity 90, member A26 |
| chr1_+_201755452 | 0.03 |
ENST00000438083.1
|
NAV1
|
neuron navigator 1 |
| chr7_-_93520191 | 0.03 |
ENST00000545378.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr10_+_90660832 | 0.03 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr7_+_92158083 | 0.03 |
ENST00000265732.5
ENST00000481551.1 ENST00000496410.1 |
RBM48
|
RNA binding motif protein 48 |
| chr13_+_49822041 | 0.03 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr9_-_80437915 | 0.03 |
ENST00000397476.3
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr20_-_36793663 | 0.03 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chrX_-_131353461 | 0.03 |
ENST00000370874.1
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr12_-_122884553 | 0.03 |
ENST00000535290.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr22_+_38035459 | 0.03 |
ENST00000357436.4
|
SH3BP1
|
SH3-domain binding protein 1 |
| chr6_-_160209471 | 0.03 |
ENST00000539948.1
|
TCP1
|
t-complex 1 |
| chr4_-_36246060 | 0.03 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr10_+_180643 | 0.02 |
ENST00000509513.2
ENST00000397959.3 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr19_+_55417499 | 0.02 |
ENST00000291890.4
ENST00000447255.1 ENST00000598576.1 ENST00000594765.1 |
NCR1
|
natural cytotoxicity triggering receptor 1 |
| chr17_-_39677971 | 0.02 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr5_-_156772729 | 0.02 |
ENST00000312349.4
|
FNDC9
|
fibronectin type III domain containing 9 |
| chr11_-_18063973 | 0.02 |
ENST00000528338.1
|
TPH1
|
tryptophan hydroxylase 1 |
| chr19_-_11688951 | 0.02 |
ENST00000589792.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr19_-_11688500 | 0.02 |
ENST00000433365.2
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr12_+_53689309 | 0.02 |
ENST00000351500.3
ENST00000550846.1 ENST00000334478.4 ENST00000549759.1 |
PFDN5
|
prefoldin subunit 5 |
| chr1_+_100503643 | 0.02 |
ENST00000370152.3
|
HIAT1
|
hippocampus abundant transcript 1 |
| chr4_-_168155417 | 0.02 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr3_-_72149553 | 0.02 |
ENST00000468646.2
ENST00000464271.1 |
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr14_+_101359265 | 0.02 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr2_+_173600514 | 0.02 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr16_+_66914264 | 0.02 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr6_+_2988199 | 0.02 |
ENST00000450238.1
ENST00000445000.1 ENST00000426637.1 |
LINC01011
NQO2
|
long intergenic non-protein coding RNA 1011 NAD(P)H dehydrogenase, quinone 2 |
| chr5_-_156772568 | 0.02 |
ENST00000520782.1
|
FNDC9
|
fibronectin type III domain containing 9 |
| chr11_+_63273811 | 0.02 |
ENST00000340246.5
|
LGALS12
|
lectin, galactoside-binding, soluble, 12 |
| chr14_-_21270995 | 0.02 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr13_+_46039037 | 0.02 |
ENST00000349995.5
|
COG3
|
component of oligomeric golgi complex 3 |
| chr7_-_3198204 | 0.02 |
ENST00000402115.1
ENST00000403226.1 |
AC091801.1
|
LOC392621; Uncharacterized protein |
| chr12_-_58329888 | 0.01 |
ENST00000546580.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr3_-_50336548 | 0.01 |
ENST00000450489.1
ENST00000513170.1 ENST00000450982.1 |
NAT6
HYAL3
|
N-acetyltransferase 6 (GCN5-related) hyaluronoglucosaminidase 3 |
| chr5_+_148443049 | 0.01 |
ENST00000515304.1
ENST00000507318.1 |
CTC-529P8.1
|
CTC-529P8.1 |
| chr14_+_67707826 | 0.01 |
ENST00000261681.4
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr6_+_26158343 | 0.01 |
ENST00000377777.4
ENST00000289316.2 |
HIST1H2BD
|
histone cluster 1, H2bd |
| chr1_-_32264356 | 0.01 |
ENST00000452755.2
|
SPOCD1
|
SPOC domain containing 1 |
| chr11_-_104905840 | 0.01 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr6_-_31782813 | 0.01 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr17_-_61781750 | 0.01 |
ENST00000582026.1
|
STRADA
|
STE20-related kinase adaptor alpha |
| chr2_+_128405655 | 0.01 |
ENST00000423019.1
|
GPR17
|
G protein-coupled receptor 17 |
| chr10_+_23216944 | 0.01 |
ENST00000298032.5
ENST00000409983.3 ENST00000409049.3 |
ARMC3
|
armadillo repeat containing 3 |
| chrX_+_140096761 | 0.01 |
ENST00000370530.1
|
SPANXB1
|
SPANX family, member B1 |
| chr15_+_93749295 | 0.01 |
ENST00000599897.1
|
AC112693.2
|
AC112693.2 |
| chr14_+_76618242 | 0.01 |
ENST00000557542.1
ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L
|
G patch domain containing 2-like |
| chr17_+_11501748 | 0.01 |
ENST00000262442.4
ENST00000579828.1 |
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr11_+_63273547 | 0.01 |
ENST00000255684.5
ENST00000394618.3 |
LGALS12
|
lectin, galactoside-binding, soluble, 12 |
| chr1_+_32757668 | 0.01 |
ENST00000373548.3
|
HDAC1
|
histone deacetylase 1 |
| chr13_+_42712178 | 0.01 |
ENST00000536612.1
|
DGKH
|
diacylglycerol kinase, eta |
| chr14_-_23877474 | 0.01 |
ENST00000405093.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr4_-_140477910 | 0.01 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr8_-_141728444 | 0.01 |
ENST00000521562.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr3_-_157217328 | 0.01 |
ENST00000392832.2
ENST00000543418.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr7_+_134430212 | 0.01 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr4_-_140477353 | 0.01 |
ENST00000406354.1
ENST00000506866.2 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr19_+_44716768 | 0.01 |
ENST00000586048.1
|
ZNF227
|
zinc finger protein 227 |
| chr11_-_74408739 | 0.00 |
ENST00000529912.1
|
CHRDL2
|
chordin-like 2 |
| chr4_-_87028478 | 0.00 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr2_+_166152283 | 0.00 |
ENST00000375427.2
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr19_+_44716678 | 0.00 |
ENST00000586228.1
ENST00000588219.1 ENST00000313040.7 ENST00000589707.1 ENST00000588394.1 ENST00000589005.1 |
ZNF227
|
zinc finger protein 227 |
| chr13_-_46716969 | 0.00 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr15_-_55563072 | 0.00 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr6_-_170599561 | 0.00 |
ENST00000366756.3
|
DLL1
|
delta-like 1 (Drosophila) |
| chr11_-_68611721 | 0.00 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chr3_+_49236910 | 0.00 |
ENST00000452691.2
ENST00000366429.2 |
CCDC36
|
coiled-coil domain containing 36 |
| chr11_-_119252359 | 0.00 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr10_-_98273668 | 0.00 |
ENST00000357947.3
|
TLL2
|
tolloid-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB12
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.2 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |