Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
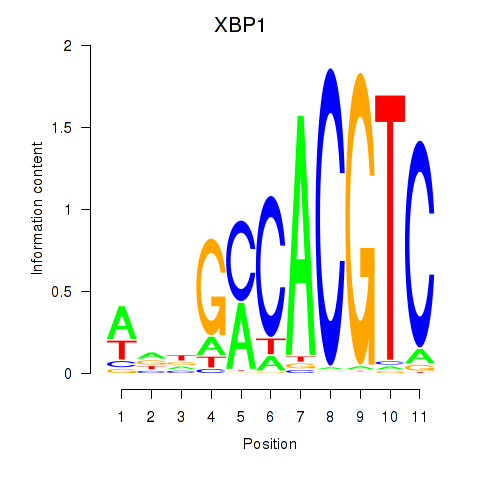
Results for XBP1
Z-value: 0.84
Transcription factors associated with XBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
XBP1
|
ENSG00000100219.12 | X-box binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| XBP1 | hg19_v2_chr22_-_29196511_29196541 | -0.48 | 3.3e-01 | Click! |
Activity profile of XBP1 motif
Sorted Z-values of XBP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_50031547 | 2.17 |
ENST00000597801.1
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr20_-_62199427 | 0.73 |
ENST00000427522.2
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr22_+_46476192 | 0.73 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr10_-_18948208 | 0.67 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr1_+_92495528 | 0.63 |
ENST00000370383.4
|
EPHX4
|
epoxide hydrolase 4 |
| chr5_+_119799927 | 0.59 |
ENST00000407149.2
ENST00000379551.2 |
PRR16
|
proline rich 16 |
| chr11_+_17298522 | 0.58 |
ENST00000529313.1
|
NUCB2
|
nucleobindin 2 |
| chr11_+_17298297 | 0.51 |
ENST00000529010.1
|
NUCB2
|
nucleobindin 2 |
| chr9_-_34523027 | 0.48 |
ENST00000399775.2
|
ENHO
|
energy homeostasis associated |
| chr22_+_38864041 | 0.46 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr7_+_72848092 | 0.45 |
ENST00000344575.3
|
FZD9
|
frizzled family receptor 9 |
| chr4_+_76649797 | 0.44 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr3_-_57583052 | 0.44 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr11_+_17298255 | 0.43 |
ENST00000531172.1
ENST00000533738.2 ENST00000323688.6 |
NUCB2
|
nucleobindin 2 |
| chr9_+_126777676 | 0.39 |
ENST00000488674.2
|
LHX2
|
LIM homeobox 2 |
| chr10_-_18948156 | 0.38 |
ENST00000414939.1
ENST00000449529.1 ENST00000456217.1 ENST00000444660.1 |
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr19_+_29704142 | 0.37 |
ENST00000587859.1
ENST00000590607.1 |
CTB-32O4.2
|
CTB-32O4.2 |
| chr3_+_132379154 | 0.37 |
ENST00000468022.1
ENST00000473651.1 ENST00000494238.2 |
UBA5
|
ubiquitin-like modifier activating enzyme 5 |
| chr2_-_38604398 | 0.35 |
ENST00000443098.1
ENST00000449130.1 ENST00000378954.4 ENST00000539122.1 ENST00000419554.2 ENST00000451483.1 ENST00000406122.1 |
ATL2
|
atlastin GTPase 2 |
| chr14_+_69951457 | 0.35 |
ENST00000322564.7
|
PLEKHD1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr4_+_76649753 | 0.34 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr21_-_18985230 | 0.34 |
ENST00000457956.1
ENST00000348354.6 |
BTG3
|
BTG family, member 3 |
| chr2_+_220408724 | 0.33 |
ENST00000421791.1
ENST00000373883.3 ENST00000451952.1 |
TMEM198
|
transmembrane protein 198 |
| chr13_-_52027134 | 0.31 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr8_+_104384616 | 0.30 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr22_+_45148432 | 0.28 |
ENST00000389774.2
ENST00000396119.2 ENST00000336963.4 ENST00000356099.6 ENST00000412433.1 |
ARHGAP8
|
Rho GTPase activating protein 8 |
| chr17_-_8059638 | 0.28 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr6_-_27841289 | 0.28 |
ENST00000355981.2
|
HIST1H4L
|
histone cluster 1, H4l |
| chr21_-_18985158 | 0.27 |
ENST00000339775.6
|
BTG3
|
BTG family, member 3 |
| chr5_+_154238149 | 0.27 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr20_-_62493217 | 0.26 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135 |
| chr4_+_128982490 | 0.26 |
ENST00000394288.3
ENST00000432347.2 ENST00000264584.5 ENST00000441387.1 ENST00000427266.1 ENST00000354456.3 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr2_-_182521699 | 0.26 |
ENST00000374969.2
ENST00000339098.5 ENST00000374970.2 |
CERKL
|
ceramide kinase-like |
| chr4_+_26322409 | 0.25 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chrX_+_134654540 | 0.25 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr11_+_17298543 | 0.25 |
ENST00000533926.1
|
NUCB2
|
nucleobindin 2 |
| chr1_+_222791417 | 0.25 |
ENST00000344922.5
ENST00000344441.6 ENST00000344507.1 |
MIA3
|
melanoma inhibitory activity family, member 3 |
| chr1_+_6845384 | 0.25 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr10_+_18948311 | 0.25 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B |
| chr7_-_30066233 | 0.24 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr1_-_22109484 | 0.24 |
ENST00000529637.1
|
USP48
|
ubiquitin specific peptidase 48 |
| chr4_-_76649546 | 0.23 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr6_-_108279369 | 0.23 |
ENST00000369002.4
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr6_+_83903061 | 0.23 |
ENST00000369724.4
ENST00000539997.1 |
RWDD2A
|
RWD domain containing 2A |
| chr5_-_115177496 | 0.22 |
ENST00000274459.4
ENST00000509910.1 |
ATG12
|
autophagy related 12 |
| chr1_-_22109682 | 0.22 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr7_+_116593953 | 0.21 |
ENST00000397750.3
|
ST7-OT4
|
ST7 overlapping transcript 4 |
| chr3_-_149688502 | 0.20 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chr5_+_154238042 | 0.20 |
ENST00000519211.1
ENST00000522458.1 ENST00000519903.1 ENST00000521450.1 ENST00000403027.2 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr1_-_155145721 | 0.20 |
ENST00000295682.4
|
KRTCAP2
|
keratinocyte associated protein 2 |
| chr4_+_128982416 | 0.20 |
ENST00000326639.6
|
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr2_+_242254753 | 0.20 |
ENST00000428524.1
ENST00000445030.1 ENST00000407017.1 |
SEPT2
|
septin 2 |
| chr11_+_32605350 | 0.20 |
ENST00000531120.1
ENST00000524896.1 ENST00000323213.5 |
EIF3M
|
eukaryotic translation initiation factor 3, subunit M |
| chr2_-_170430277 | 0.19 |
ENST00000438035.1
ENST00000453929.2 |
FASTKD1
|
FAST kinase domains 1 |
| chr8_-_124054362 | 0.19 |
ENST00000405944.3
|
DERL1
|
derlin 1 |
| chr2_-_33824382 | 0.19 |
ENST00000238823.8
|
FAM98A
|
family with sequence similarity 98, member A |
| chr2_+_64751433 | 0.19 |
ENST00000238856.4
ENST00000422803.1 ENST00000238855.7 |
AFTPH
|
aftiphilin |
| chr3_-_57583130 | 0.18 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr13_-_45915221 | 0.18 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr5_+_154237778 | 0.18 |
ENST00000523698.1
ENST00000517876.1 ENST00000520472.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr1_+_101361782 | 0.18 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr1_+_87170577 | 0.17 |
ENST00000482504.1
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1 |
| chr22_-_43253189 | 0.17 |
ENST00000437119.2
ENST00000429508.2 ENST00000454099.1 ENST00000263245.5 |
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr3_-_132378919 | 0.17 |
ENST00000355458.3
|
ACAD11
|
acyl-CoA dehydrogenase family, member 11 |
| chr19_+_1491144 | 0.17 |
ENST00000233596.3
|
REEP6
|
receptor accessory protein 6 |
| chr16_+_777118 | 0.17 |
ENST00000562141.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr9_+_139249228 | 0.17 |
ENST00000392944.1
|
GPSM1
|
G-protein signaling modulator 1 |
| chr3_+_132378741 | 0.16 |
ENST00000493720.2
|
UBA5
|
ubiquitin-like modifier activating enzyme 5 |
| chr7_-_27183263 | 0.16 |
ENST00000222726.3
|
HOXA5
|
homeobox A5 |
| chr8_-_124054587 | 0.16 |
ENST00000259512.4
|
DERL1
|
derlin 1 |
| chr4_+_141445311 | 0.16 |
ENST00000323570.3
ENST00000511887.2 |
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr7_+_128379346 | 0.16 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr5_+_154238096 | 0.15 |
ENST00000517568.1
ENST00000524105.1 ENST00000285896.6 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr5_+_9546306 | 0.15 |
ENST00000508179.1
|
SNHG18
|
small nucleolar RNA host gene 18 (non-protein coding) |
| chr12_-_99038732 | 0.15 |
ENST00000393042.3
ENST00000420861.1 ENST00000299157.4 ENST00000342502.2 |
IKBIP
|
IKBKB interacting protein |
| chr12_-_28123206 | 0.15 |
ENST00000542963.1
ENST00000535992.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr5_-_108745689 | 0.14 |
ENST00000361189.2
|
PJA2
|
praja ring finger 2, E3 ubiquitin protein ligase |
| chr13_-_52026730 | 0.14 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr4_+_141445333 | 0.14 |
ENST00000507667.1
|
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr14_-_92506371 | 0.14 |
ENST00000267622.4
|
TRIP11
|
thyroid hormone receptor interactor 11 |
| chr6_-_109702885 | 0.14 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr3_-_149688896 | 0.14 |
ENST00000239940.7
|
PFN2
|
profilin 2 |
| chr7_-_12443501 | 0.14 |
ENST00000275358.3
|
VWDE
|
von Willebrand factor D and EGF domains |
| chr9_+_37079888 | 0.13 |
ENST00000429493.1
ENST00000593237.1 ENST00000588557.1 ENST00000430809.1 ENST00000592157.1 |
RP11-220I1.1
|
RP11-220I1.1 |
| chr5_+_9546376 | 0.13 |
ENST00000509788.1
|
SNHG18
|
small nucleolar RNA host gene 18 (non-protein coding) |
| chr18_-_44775554 | 0.13 |
ENST00000425639.1
ENST00000400404.1 |
SKOR2
|
SKI family transcriptional corepressor 2 |
| chr15_-_80695917 | 0.13 |
ENST00000559008.1
|
RP11-210M15.2
|
Uncharacterized protein |
| chr7_+_97840739 | 0.13 |
ENST00000609256.1
|
BHLHA15
|
basic helix-loop-helix family, member a15 |
| chr2_+_242254679 | 0.13 |
ENST00000428282.1
ENST00000360051.3 |
SEPT2
|
septin 2 |
| chr6_-_83902933 | 0.12 |
ENST00000512866.1
ENST00000510258.1 ENST00000503094.1 ENST00000283977.4 ENST00000513973.1 ENST00000508748.1 |
PGM3
|
phosphoglucomutase 3 |
| chr20_+_32951041 | 0.12 |
ENST00000374864.4
|
ITCH
|
itchy E3 ubiquitin protein ligase |
| chr6_-_7313381 | 0.12 |
ENST00000489567.1
ENST00000479365.1 ENST00000462112.1 ENST00000397511.2 ENST00000534851.1 ENST00000474597.1 ENST00000244763.4 |
SSR1
|
signal sequence receptor, alpha |
| chr2_-_20251744 | 0.12 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr5_-_176827631 | 0.12 |
ENST00000358571.2
|
PFN3
|
profilin 3 |
| chr1_-_144932014 | 0.12 |
ENST00000529945.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr17_-_42276574 | 0.12 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr5_+_112196919 | 0.12 |
ENST00000505459.1
ENST00000282999.3 ENST00000515463.1 |
SRP19
|
signal recognition particle 19kDa |
| chr13_+_46039037 | 0.12 |
ENST00000349995.5
|
COG3
|
component of oligomeric golgi complex 3 |
| chr6_-_90062543 | 0.12 |
ENST00000435041.2
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr6_+_132129151 | 0.12 |
ENST00000360971.2
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr1_-_119683251 | 0.12 |
ENST00000369426.5
ENST00000235521.4 |
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr1_-_119682812 | 0.12 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr12_+_107349497 | 0.11 |
ENST00000548125.1
ENST00000280756.4 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr12_-_28122980 | 0.11 |
ENST00000395868.3
ENST00000534890.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr5_+_121187625 | 0.11 |
ENST00000321339.1
|
FTMT
|
ferritin mitochondrial |
| chr3_-_57583185 | 0.11 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr14_+_31091511 | 0.11 |
ENST00000544052.2
ENST00000421551.3 ENST00000541123.1 ENST00000557076.1 ENST00000553693.1 ENST00000396629.2 |
SCFD1
|
sec1 family domain containing 1 |
| chr20_+_57467204 | 0.11 |
ENST00000603546.1
|
GNAS
|
GNAS complex locus |
| chr14_+_96342729 | 0.11 |
ENST00000504119.1
|
LINC00617
|
long intergenic non-protein coding RNA 617 |
| chr6_+_116692102 | 0.10 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chr2_+_242254507 | 0.10 |
ENST00000391973.2
|
SEPT2
|
septin 2 |
| chr2_-_99952769 | 0.10 |
ENST00000409434.1
ENST00000434323.1 ENST00000264255.3 |
TXNDC9
|
thioredoxin domain containing 9 |
| chr3_-_132378897 | 0.10 |
ENST00000545291.1
|
ACAD11
|
acyl-CoA dehydrogenase family, member 11 |
| chr7_-_6523688 | 0.09 |
ENST00000490996.1
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr2_-_170430366 | 0.09 |
ENST00000453153.2
ENST00000445210.1 |
FASTKD1
|
FAST kinase domains 1 |
| chr3_-_28390120 | 0.09 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr5_-_9546180 | 0.09 |
ENST00000382496.5
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr2_-_33824336 | 0.08 |
ENST00000431950.1
ENST00000403368.1 ENST00000441530.2 |
FAM98A
|
family with sequence similarity 98, member A |
| chr14_+_39736582 | 0.08 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr2_+_171785824 | 0.08 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr17_+_72199721 | 0.08 |
ENST00000439590.2
ENST00000311111.6 ENST00000584577.1 ENST00000534490.1 ENST00000528433.2 ENST00000533498.1 |
RPL38
|
ribosomal protein L38 |
| chr17_+_15848231 | 0.08 |
ENST00000304222.2
|
ADORA2B
|
adenosine A2b receptor |
| chr10_+_64893039 | 0.08 |
ENST00000277746.6
ENST00000435510.2 |
NRBF2
|
nuclear receptor binding factor 2 |
| chr3_-_139108475 | 0.08 |
ENST00000515006.1
ENST00000513274.1 ENST00000514508.1 ENST00000507777.1 ENST00000512153.1 ENST00000333188.5 |
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr12_-_106641728 | 0.08 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr7_+_128379449 | 0.07 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr4_+_128982430 | 0.07 |
ENST00000512292.1
ENST00000508819.1 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr6_+_63921351 | 0.07 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr19_-_54618650 | 0.07 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr4_-_83351005 | 0.07 |
ENST00000295470.5
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr6_-_111927062 | 0.07 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr14_-_50087312 | 0.07 |
ENST00000298289.6
|
RPL36AL
|
ribosomal protein L36a-like |
| chr1_-_87380002 | 0.07 |
ENST00000331835.5
|
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr13_-_31040060 | 0.07 |
ENST00000326004.4
ENST00000341423.5 |
HMGB1
|
high mobility group box 1 |
| chr11_-_118927880 | 0.07 |
ENST00000527038.2
|
HYOU1
|
hypoxia up-regulated 1 |
| chr9_+_114393581 | 0.07 |
ENST00000313525.3
|
DNAJC25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr16_-_30546275 | 0.06 |
ENST00000568028.1
|
ZNF747
|
zinc finger protein 747 |
| chr16_+_2083265 | 0.06 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr11_+_118443098 | 0.06 |
ENST00000392859.3
ENST00000359415.4 ENST00000534182.2 ENST00000264028.4 |
ARCN1
|
archain 1 |
| chr11_-_118927561 | 0.06 |
ENST00000530473.1
|
HYOU1
|
hypoxia up-regulated 1 |
| chr3_+_105086056 | 0.06 |
ENST00000472644.2
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr4_+_190992087 | 0.06 |
ENST00000553598.1
|
DUX4L7
|
double homeobox 4 like 7 |
| chr10_+_32735030 | 0.05 |
ENST00000277657.6
ENST00000362006.5 |
CCDC7
|
coiled-coil domain containing 7 |
| chr2_+_64681219 | 0.05 |
ENST00000238875.5
|
LGALSL
|
lectin, galactoside-binding-like |
| chr1_+_6845497 | 0.05 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr11_-_100999775 | 0.05 |
ENST00000263463.5
|
PGR
|
progesterone receptor |
| chr18_-_46987000 | 0.05 |
ENST00000442713.2
ENST00000269445.6 |
DYM
|
dymeclin |
| chr4_+_165675197 | 0.05 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr3_-_28390298 | 0.05 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr20_-_17662705 | 0.05 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr6_+_7107999 | 0.05 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr17_-_42991062 | 0.04 |
ENST00000587997.1
|
GFAP
|
glial fibrillary acidic protein |
| chr17_+_8339164 | 0.04 |
ENST00000582665.1
ENST00000334527.7 ENST00000299734.7 |
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr2_-_15701422 | 0.04 |
ENST00000441750.1
ENST00000281513.5 |
NBAS
|
neuroblastoma amplified sequence |
| chr11_+_64851666 | 0.04 |
ENST00000525509.1
ENST00000294258.3 ENST00000526334.1 |
ZFPL1
|
zinc finger protein-like 1 |
| chr20_-_33680588 | 0.04 |
ENST00000451813.2
ENST00000432634.2 |
TRPC4AP
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr11_-_118972575 | 0.04 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr19_-_49015050 | 0.04 |
ENST00000600059.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr1_-_87379785 | 0.04 |
ENST00000401030.3
ENST00000370554.1 |
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr17_+_8339189 | 0.04 |
ENST00000585098.1
ENST00000380025.4 ENST00000402554.3 ENST00000584866.1 ENST00000582490.1 |
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr11_-_118927735 | 0.04 |
ENST00000526656.1
|
HYOU1
|
hypoxia up-regulated 1 |
| chr14_+_39736299 | 0.04 |
ENST00000341502.5
ENST00000396158.2 ENST00000280083.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr10_+_89419370 | 0.03 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr9_-_112260531 | 0.03 |
ENST00000374541.2
ENST00000262539.3 |
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr9_+_101984577 | 0.03 |
ENST00000223641.4
|
SEC61B
|
Sec61 beta subunit |
| chr1_-_144932316 | 0.03 |
ENST00000313431.9
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr4_+_108746282 | 0.03 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr5_+_126853301 | 0.03 |
ENST00000296666.8
ENST00000442138.2 ENST00000512635.2 |
PRRC1
|
proline-rich coiled-coil 1 |
| chr9_+_15552867 | 0.03 |
ENST00000535968.1
|
CCDC171
|
coiled-coil domain containing 171 |
| chr4_+_40058411 | 0.03 |
ENST00000261435.6
ENST00000515550.1 |
N4BP2
|
NEDD4 binding protein 2 |
| chr4_-_119757322 | 0.03 |
ENST00000379735.5
|
SEC24D
|
SEC24 family member D |
| chr8_-_81083890 | 0.03 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr19_-_54619006 | 0.03 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr8_+_56014949 | 0.03 |
ENST00000327381.6
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4 |
| chr2_-_214017151 | 0.03 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr20_+_32951070 | 0.03 |
ENST00000535650.1
ENST00000262650.6 |
ITCH
|
itchy E3 ubiquitin protein ligase |
| chr4_-_83350580 | 0.02 |
ENST00000349655.4
ENST00000602300.1 |
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr6_-_43596899 | 0.02 |
ENST00000307126.5
ENST00000452781.1 |
GTPBP2
|
GTP binding protein 2 |
| chr1_+_249200395 | 0.02 |
ENST00000355360.4
ENST00000329291.5 ENST00000539153.1 |
PGBD2
|
piggyBac transposable element derived 2 |
| chr14_-_24664540 | 0.02 |
ENST00000530563.1
ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1
|
transmembrane 9 superfamily member 1 |
| chr18_-_47017956 | 0.02 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr1_+_110527308 | 0.02 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr3_-_156272924 | 0.02 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr7_+_6522922 | 0.02 |
ENST00000601673.1
|
FLJ20306
|
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein |
| chr20_-_17662878 | 0.01 |
ENST00000377813.1
ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1
|
ribosome binding protein 1 |
| chr2_-_88927092 | 0.01 |
ENST00000303236.3
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr18_+_44497455 | 0.01 |
ENST00000592005.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr19_+_7701985 | 0.01 |
ENST00000595950.1
ENST00000441779.2 ENST00000221283.5 ENST00000414284.2 |
STXBP2
|
syntaxin binding protein 2 |
| chr13_+_26042960 | 0.01 |
ENST00000255283.8
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2 |
| chr1_+_207669573 | 0.00 |
ENST00000400960.2
ENST00000534202.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr8_-_124054484 | 0.00 |
ENST00000419562.2
|
DERL1
|
derlin 1 |
| chr11_-_118927816 | 0.00 |
ENST00000534233.1
ENST00000532752.1 ENST00000525859.1 ENST00000404233.3 ENST00000532421.1 ENST00000543287.1 ENST00000527310.2 ENST00000529972.1 |
HYOU1
|
hypoxia up-regulated 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of XBP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.5 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.5 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.4 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.2 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:1903382 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.0 | 0.2 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.3 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.2 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.4 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0002880 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.4 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.1 | 0.3 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.1 | 0.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |