Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
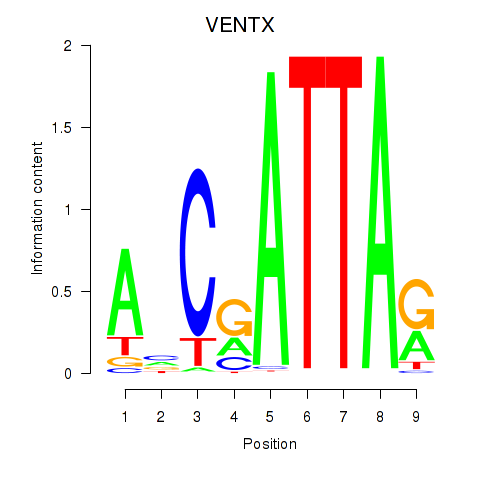
Results for VENTX
Z-value: 0.51
Transcription factors associated with VENTX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VENTX
|
ENSG00000151650.7 | VENT homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VENTX | hg19_v2_chr10_+_135050908_135050908 | 0.02 | 9.7e-01 | Click! |
Activity profile of VENTX motif
Sorted Z-values of VENTX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_110958124 | 0.78 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr17_+_47448102 | 0.36 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr12_-_86650077 | 0.33 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr18_+_20714525 | 0.31 |
ENST00000400473.2
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr12_+_107712173 | 0.30 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr17_-_46716647 | 0.28 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr4_-_84035905 | 0.28 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr2_+_45168875 | 0.26 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr12_-_120241187 | 0.24 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr4_+_169418255 | 0.23 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr11_-_111649074 | 0.23 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr6_-_138820624 | 0.23 |
ENST00000343505.5
|
NHSL1
|
NHS-like 1 |
| chr11_-_72070206 | 0.21 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr3_+_167582561 | 0.20 |
ENST00000463642.1
ENST00000464514.1 |
RP11-298O21.6
RP11-298O21.7
|
RP11-298O21.6 RP11-298O21.7 |
| chr1_+_62439037 | 0.20 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr16_+_70207686 | 0.20 |
ENST00000541793.2
ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18, member C |
| chr6_-_90529418 | 0.19 |
ENST00000439638.1
ENST00000369393.3 ENST00000428876.1 |
MDN1
|
MDN1, midasin homolog (yeast) |
| chr18_+_13465008 | 0.18 |
ENST00000593236.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr14_+_32798547 | 0.18 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr11_-_111649015 | 0.18 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr7_+_139025875 | 0.18 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr11_+_46722368 | 0.17 |
ENST00000311764.2
|
ZNF408
|
zinc finger protein 408 |
| chr1_+_174417095 | 0.17 |
ENST00000367685.2
|
GPR52
|
G protein-coupled receptor 52 |
| chr1_-_232651312 | 0.16 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr1_-_67600639 | 0.16 |
ENST00000544837.1
ENST00000603691.1 |
C1orf141
|
chromosome 1 open reading frame 141 |
| chr9_+_116225999 | 0.16 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chr11_-_64684672 | 0.15 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr19_-_46088068 | 0.15 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chrX_-_153602991 | 0.15 |
ENST00000369850.3
ENST00000422373.1 |
FLNA
|
filamin A, alpha |
| chr1_+_153746683 | 0.15 |
ENST00000271857.2
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr14_+_65182395 | 0.14 |
ENST00000554088.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr4_+_129732467 | 0.14 |
ENST00000413543.2
|
PHF17
|
jade family PHD finger 1 |
| chr12_+_25205446 | 0.14 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr19_+_48949087 | 0.14 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr22_+_39101728 | 0.14 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr20_-_25371412 | 0.14 |
ENST00000339157.5
ENST00000376542.3 |
ABHD12
|
abhydrolase domain containing 12 |
| chr1_+_203651937 | 0.14 |
ENST00000341360.2
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr16_+_14280742 | 0.14 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr1_-_23886285 | 0.13 |
ENST00000374561.5
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr4_-_84035868 | 0.13 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr4_+_169418195 | 0.13 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr6_+_114178512 | 0.12 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr1_+_28527070 | 0.12 |
ENST00000596102.1
|
AL353354.2
|
AL353354.2 |
| chr19_+_48949030 | 0.12 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr9_+_117904097 | 0.12 |
ENST00000374016.1
|
DEC1
|
deleted in esophageal cancer 1 |
| chr9_+_2159850 | 0.12 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_153747746 | 0.12 |
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr12_-_51611477 | 0.12 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr17_+_17685422 | 0.11 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr4_+_129732419 | 0.11 |
ENST00000510308.1
|
PHF17
|
jade family PHD finger 1 |
| chr1_+_43291220 | 0.11 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr8_-_116681123 | 0.11 |
ENST00000519674.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr4_-_122148620 | 0.11 |
ENST00000509841.1
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr11_-_3859089 | 0.11 |
ENST00000396979.1
|
RHOG
|
ras homolog family member G |
| chr18_-_74207146 | 0.11 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr1_+_168250194 | 0.11 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr6_-_161695074 | 0.11 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr10_-_49812997 | 0.10 |
ENST00000417912.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr3_-_27764190 | 0.10 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr6_+_37897735 | 0.10 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr10_+_24738355 | 0.10 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr2_+_226265364 | 0.10 |
ENST00000272907.6
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chrX_+_135730297 | 0.10 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr2_-_224467002 | 0.10 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr12_+_25205568 | 0.10 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr12_+_52695617 | 0.10 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr20_+_42136308 | 0.10 |
ENST00000434666.1
ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr16_+_73420942 | 0.09 |
ENST00000554640.1
ENST00000562661.1 ENST00000561875.1 |
RP11-140I24.1
|
RP11-140I24.1 |
| chr14_+_65171099 | 0.09 |
ENST00000247226.7
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr12_+_25205666 | 0.09 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr18_-_24237339 | 0.08 |
ENST00000580191.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr14_+_65171315 | 0.08 |
ENST00000394691.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr10_-_28571015 | 0.08 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr12_-_86650045 | 0.08 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr14_+_65453432 | 0.08 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr11_+_7618413 | 0.08 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr11_+_123430948 | 0.07 |
ENST00000529432.1
ENST00000534764.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr14_-_27066636 | 0.07 |
ENST00000267422.7
ENST00000344429.5 ENST00000574031.1 ENST00000465357.2 ENST00000547619.1 |
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr3_+_171561127 | 0.07 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr11_+_64018955 | 0.07 |
ENST00000279230.6
ENST00000540288.1 ENST00000325234.5 |
PLCB3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr3_+_186383741 | 0.07 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chrX_+_55478538 | 0.07 |
ENST00000342972.1
|
MAGEH1
|
melanoma antigen family H, 1 |
| chr2_+_135596106 | 0.07 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr3_+_32859510 | 0.07 |
ENST00000383763.5
|
TRIM71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr12_-_89746264 | 0.06 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr1_+_62417957 | 0.06 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr12_-_122018114 | 0.06 |
ENST00000539394.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr17_+_39845134 | 0.06 |
ENST00000591776.1
ENST00000469257.1 |
EIF1
|
eukaryotic translation initiation factor 1 |
| chr12_+_70219052 | 0.06 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chrX_+_135730373 | 0.06 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr3_-_9834463 | 0.06 |
ENST00000439043.1
|
TADA3
|
transcriptional adaptor 3 |
| chr13_-_67802549 | 0.05 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr16_+_14280564 | 0.05 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr6_+_101846664 | 0.05 |
ENST00000421544.1
ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2
|
glutamate receptor, ionotropic, kainate 2 |
| chr12_-_2113583 | 0.05 |
ENST00000397173.4
ENST00000280665.6 |
DCP1B
|
decapping mRNA 1B |
| chr4_+_123161120 | 0.05 |
ENST00000446180.1
|
KIAA1109
|
KIAA1109 |
| chr6_+_34204642 | 0.05 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr15_+_54305101 | 0.05 |
ENST00000260323.11
ENST00000545554.1 ENST00000537900.1 |
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr5_-_41794313 | 0.05 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr2_-_145188137 | 0.05 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr4_+_169013666 | 0.05 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr16_+_69985083 | 0.05 |
ENST00000288040.6
ENST00000449317.2 |
CLEC18A
|
C-type lectin domain family 18, member A |
| chr10_-_49813090 | 0.05 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr7_+_141408153 | 0.05 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr8_-_116681221 | 0.05 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr7_+_129984630 | 0.05 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr1_+_162351503 | 0.05 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr3_+_9834227 | 0.04 |
ENST00000287613.7
ENST00000397261.3 |
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa |
| chr7_+_18535321 | 0.04 |
ENST00000413380.1
ENST00000430454.1 |
HDAC9
|
histone deacetylase 9 |
| chr6_+_152011628 | 0.04 |
ENST00000404742.1
ENST00000440973.1 |
ESR1
|
estrogen receptor 1 |
| chr9_+_133539981 | 0.04 |
ENST00000253008.2
|
PRDM12
|
PR domain containing 12 |
| chr7_+_18535346 | 0.04 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chr5_-_115890554 | 0.04 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr18_-_21977748 | 0.04 |
ENST00000399441.4
ENST00000319481.3 |
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr6_+_167704798 | 0.04 |
ENST00000230256.3
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr3_+_185303962 | 0.04 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr6_+_126221034 | 0.04 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr17_-_47045949 | 0.04 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr4_-_168155169 | 0.04 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr6_+_167704838 | 0.03 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr2_+_177015122 | 0.03 |
ENST00000468418.3
|
HOXD3
|
homeobox D3 |
| chr11_-_117170403 | 0.03 |
ENST00000504995.1
|
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr1_+_150337144 | 0.03 |
ENST00000539519.1
ENST00000369067.3 ENST00000369068.4 |
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr18_+_55888767 | 0.03 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr1_-_144994909 | 0.03 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr4_-_176733377 | 0.03 |
ENST00000505375.1
|
GPM6A
|
glycoprotein M6A |
| chr2_+_203776937 | 0.03 |
ENST00000402905.3
ENST00000414490.1 ENST00000431787.1 ENST00000444724.1 ENST00000414857.1 ENST00000430899.1 ENST00000445120.1 ENST00000441569.1 ENST00000432024.1 ENST00000443740.1 ENST00000414439.1 ENST00000428585.1 ENST00000545253.1 ENST00000545262.1 ENST00000447539.1 ENST00000456821.2 ENST00000434998.1 ENST00000320443.8 |
CARF
|
calcium responsive transcription factor |
| chr2_+_159825143 | 0.03 |
ENST00000454300.1
ENST00000263635.6 |
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr12_-_122017542 | 0.03 |
ENST00000446152.2
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr8_+_38831683 | 0.03 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chrX_-_21676442 | 0.03 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr2_+_87135076 | 0.03 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr3_+_9834179 | 0.03 |
ENST00000498623.2
|
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa |
| chr11_-_107729887 | 0.03 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr8_+_97773202 | 0.02 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr17_+_43239231 | 0.02 |
ENST00000591576.1
ENST00000591070.1 ENST00000592695.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chrX_+_105936982 | 0.02 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr3_-_157221380 | 0.02 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_-_88285309 | 0.02 |
ENST00000420840.2
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr1_+_40713573 | 0.02 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr8_-_109799793 | 0.02 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr8_+_97773457 | 0.02 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr2_-_224467093 | 0.02 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr1_-_178840157 | 0.02 |
ENST00000367629.1
ENST00000234816.2 |
ANGPTL1
|
angiopoietin-like 1 |
| chr6_-_66417107 | 0.02 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr14_+_32798462 | 0.02 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr17_+_59489112 | 0.02 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr6_-_161695042 | 0.02 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr1_+_186265399 | 0.02 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr12_+_126107042 | 0.02 |
ENST00000535886.1
|
TMEM132B
|
transmembrane protein 132B |
| chr17_-_29645836 | 0.02 |
ENST00000578584.1
|
CTD-2370N5.3
|
CTD-2370N5.3 |
| chr2_+_135596180 | 0.01 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr6_+_153552455 | 0.01 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr10_-_50603497 | 0.01 |
ENST00000374139.2
|
DRGX
|
dorsal root ganglia homeobox |
| chr8_+_31496809 | 0.01 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr19_-_49622348 | 0.01 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr7_-_82792215 | 0.01 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr18_-_53253000 | 0.01 |
ENST00000566514.1
|
TCF4
|
transcription factor 4 |
| chr12_-_122018346 | 0.01 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr21_+_39644395 | 0.01 |
ENST00000398934.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr5_+_43121698 | 0.00 |
ENST00000505606.2
ENST00000509634.1 ENST00000509341.1 |
ZNF131
|
zinc finger protein 131 |
| chr17_-_48785216 | 0.00 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr6_-_111927449 | 0.00 |
ENST00000368761.5
ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr3_+_156799587 | 0.00 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr5_-_156486120 | 0.00 |
ENST00000522693.1
|
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chr14_+_92789498 | 0.00 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr17_-_202579 | 0.00 |
ENST00000577079.1
ENST00000331302.7 ENST00000536489.2 |
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr1_+_41174988 | 0.00 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr4_+_37455536 | 0.00 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of VENTX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.2 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:1903248 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.0 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 0.1 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |