Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
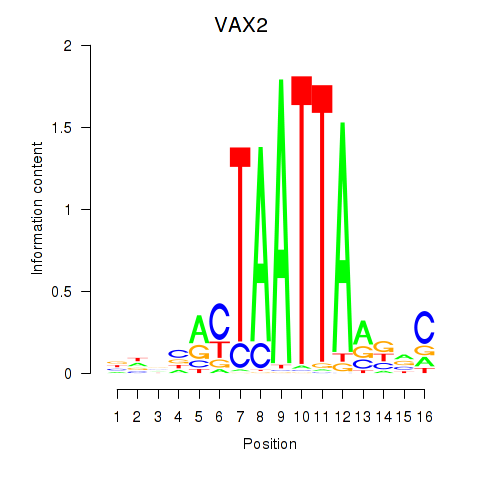
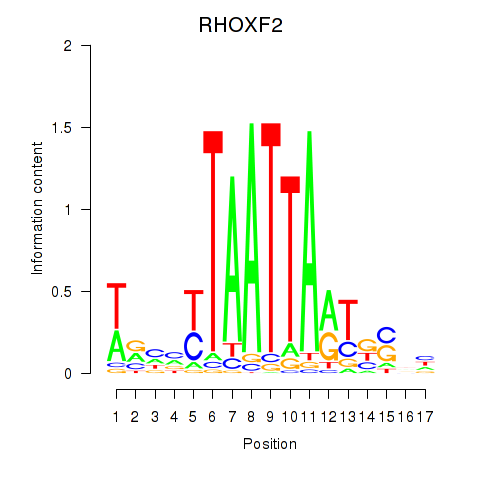
Results for VAX2_RHOXF2
Z-value: 0.45
Transcription factors associated with VAX2_RHOXF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VAX2
|
ENSG00000116035.2 | ventral anterior homeobox 2 |
|
RHOXF2
|
ENSG00000131721.4 | Rhox homeobox family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VAX2 | hg19_v2_chr2_+_71127699_71127744 | 0.58 | 2.2e-01 | Click! |
| RHOXF2 | hg19_v2_chrX_+_119292467_119292467 | -0.04 | 9.4e-01 | Click! |
Activity profile of VAX2_RHOXF2 motif
Sorted Z-values of VAX2_RHOXF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_162336686 | 0.48 |
ENST00000420220.1
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr7_-_99716914 | 0.45 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr6_+_26402517 | 0.31 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr1_+_62439037 | 0.28 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr19_-_52307357 | 0.24 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr6_+_26402465 | 0.20 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr2_-_74619152 | 0.20 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr2_-_74618964 | 0.18 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr6_-_33663474 | 0.17 |
ENST00000594414.1
|
SBP1
|
SBP1; Uncharacterized protein |
| chr17_+_4692230 | 0.17 |
ENST00000331264.7
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr12_+_78359999 | 0.16 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr4_+_169418255 | 0.15 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chrX_+_10126488 | 0.15 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr4_+_66536248 | 0.15 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr6_+_26365443 | 0.15 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr1_+_225600404 | 0.13 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr14_+_56584414 | 0.13 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr16_+_69345243 | 0.13 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr19_-_3557570 | 0.12 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr7_-_99716952 | 0.12 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr7_-_99716940 | 0.12 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr17_-_40337470 | 0.12 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr5_-_136649218 | 0.12 |
ENST00000510405.1
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr3_-_112127981 | 0.12 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr1_-_57045228 | 0.12 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr4_+_169418195 | 0.11 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_+_43238438 | 0.11 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr4_+_159727272 | 0.11 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr3_-_191000172 | 0.11 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr1_-_159832438 | 0.10 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr7_-_99717463 | 0.10 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr19_+_11071546 | 0.10 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr3_+_157154578 | 0.10 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr7_+_148936732 | 0.10 |
ENST00000335870.2
|
ZNF212
|
zinc finger protein 212 |
| chr6_+_26365387 | 0.10 |
ENST00000532865.1
ENST00000530653.1 ENST00000527417.1 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr19_-_46088068 | 0.10 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr15_+_58702742 | 0.09 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr7_-_100860851 | 0.09 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr20_+_43990576 | 0.09 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr1_+_160370344 | 0.09 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr10_+_18549645 | 0.08 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr1_+_35258592 | 0.08 |
ENST00000342280.4
ENST00000450137.1 |
GJA4
|
gap junction protein, alpha 4, 37kDa |
| chr7_+_100860949 | 0.08 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr14_+_32798547 | 0.08 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chrX_+_135730297 | 0.08 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr10_+_99205894 | 0.08 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chrX_+_135730373 | 0.07 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr10_+_6779326 | 0.07 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr17_+_40610862 | 0.07 |
ENST00000393829.2
ENST00000546249.1 ENST00000537728.1 ENST00000264649.6 ENST00000585525.1 ENST00000343619.4 ENST00000544137.1 ENST00000589727.1 ENST00000587824.1 |
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1 |
| chr1_-_201140673 | 0.07 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr11_+_71900703 | 0.06 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr6_+_3259148 | 0.06 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr11_+_33061543 | 0.06 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr20_-_33735070 | 0.06 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr9_-_5339873 | 0.06 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr14_-_20923195 | 0.06 |
ENST00000206542.4
|
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr15_+_89922345 | 0.06 |
ENST00000558982.1
|
LINC00925
|
long intergenic non-protein coding RNA 925 |
| chr12_-_118797475 | 0.05 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr1_+_54569968 | 0.05 |
ENST00000391366.1
|
AL161915.1
|
Uncharacterized protein |
| chr10_+_99205959 | 0.05 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr12_+_26348246 | 0.05 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr8_-_1922789 | 0.05 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr10_-_92681033 | 0.05 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr4_+_88571429 | 0.05 |
ENST00000339673.6
ENST00000282479.7 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr4_-_186696425 | 0.04 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_-_149900122 | 0.04 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr12_-_118796910 | 0.04 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr12_+_131438443 | 0.04 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr12_+_26348429 | 0.04 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr7_-_44122063 | 0.04 |
ENST00000335195.6
ENST00000395831.3 ENST00000414235.1 ENST00000452049.1 ENST00000242248.5 |
POLM
|
polymerase (DNA directed), mu |
| chr4_-_186696515 | 0.04 |
ENST00000456596.1
ENST00000414724.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_99717230 | 0.04 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr4_-_186696561 | 0.04 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_+_34204642 | 0.04 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr2_+_169926047 | 0.04 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chrX_+_7137475 | 0.03 |
ENST00000217961.4
|
STS
|
steroid sulfatase (microsomal), isozyme S |
| chr12_-_9760482 | 0.03 |
ENST00000229402.3
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1 |
| chr16_-_55866997 | 0.03 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chr6_+_130339710 | 0.03 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr1_+_153747746 | 0.03 |
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr8_-_91095099 | 0.03 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chrX_+_36254051 | 0.03 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr9_-_95166841 | 0.03 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr9_+_125132803 | 0.03 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr17_-_41466555 | 0.03 |
ENST00000586231.1
|
LINC00910
|
long intergenic non-protein coding RNA 910 |
| chr6_+_167536230 | 0.03 |
ENST00000341935.5
ENST00000349984.4 |
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr11_+_71900572 | 0.03 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr7_+_6655225 | 0.03 |
ENST00000457543.3
|
ZNF853
|
zinc finger protein 853 |
| chr4_-_69111401 | 0.03 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr5_+_129083772 | 0.03 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr11_-_107729887 | 0.03 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr1_-_24151903 | 0.03 |
ENST00000436439.2
ENST00000374490.3 |
HMGCL
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr18_-_67623906 | 0.02 |
ENST00000583955.1
|
CD226
|
CD226 molecule |
| chr7_+_73245193 | 0.02 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr5_+_66300446 | 0.02 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chrX_+_43515467 | 0.02 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr20_-_50722183 | 0.02 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr14_+_101359265 | 0.02 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr5_+_140557371 | 0.02 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr7_+_115862858 | 0.02 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr2_-_85555385 | 0.02 |
ENST00000377386.3
|
TGOLN2
|
trans-golgi network protein 2 |
| chr9_-_95166884 | 0.02 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr11_-_62521614 | 0.02 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr15_-_37393406 | 0.02 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr6_-_138866823 | 0.02 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chrX_-_77225135 | 0.02 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr4_-_77328458 | 0.02 |
ENST00000388914.3
ENST00000434846.2 |
CCDC158
|
coiled-coil domain containing 158 |
| chr2_-_74618907 | 0.02 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chr4_-_74486347 | 0.02 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr19_+_36632485 | 0.02 |
ENST00000586963.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr19_+_36632204 | 0.01 |
ENST00000592354.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr2_-_214016314 | 0.01 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr18_-_67624412 | 0.01 |
ENST00000580335.1
|
CD226
|
CD226 molecule |
| chr2_+_228736321 | 0.01 |
ENST00000309931.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr15_+_58430567 | 0.01 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr6_+_53883708 | 0.01 |
ENST00000514921.1
ENST00000274897.5 ENST00000370877.2 |
MLIP
|
muscular LMNA-interacting protein |
| chr4_+_88896819 | 0.01 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr16_-_28608424 | 0.01 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr14_+_74034310 | 0.01 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr18_-_67624160 | 0.01 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr14_+_103851712 | 0.01 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr4_+_151503077 | 0.01 |
ENST00000317605.4
|
MAB21L2
|
mab-21-like 2 (C. elegans) |
| chr20_+_15177480 | 0.01 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr10_-_99205607 | 0.01 |
ENST00000477692.2
ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1
|
exosome component 1 |
| chr14_+_20923350 | 0.01 |
ENST00000555414.1
ENST00000216714.3 ENST00000553681.1 ENST00000557344.1 ENST00000398030.4 ENST00000557181.1 ENST00000555839.1 ENST00000553368.1 ENST00000556054.1 ENST00000557054.1 ENST00000557592.1 ENST00000557150.1 |
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr9_+_12693336 | 0.01 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr17_+_66511540 | 0.01 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr15_-_49912944 | 0.01 |
ENST00000559905.1
|
FAM227B
|
family with sequence similarity 227, member B |
| chr5_-_115890554 | 0.01 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr19_+_12175504 | 0.01 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr14_+_32798462 | 0.00 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr6_+_26440700 | 0.00 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr9_+_125133467 | 0.00 |
ENST00000426608.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr13_-_36050819 | 0.00 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr16_+_72088376 | 0.00 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr1_+_75600567 | 0.00 |
ENST00000356261.3
|
LHX8
|
LIM homeobox 8 |
| chr15_+_49462397 | 0.00 |
ENST00000396509.2
|
GALK2
|
galactokinase 2 |
| chr2_-_198540751 | 0.00 |
ENST00000429081.1
|
RFTN2
|
raftlin family member 2 |
| chr5_-_149829244 | 0.00 |
ENST00000312037.5
|
RPS14
|
ribosomal protein S14 |
| chr17_-_71258491 | 0.00 |
ENST00000397671.1
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like |
| chr2_+_200472779 | 0.00 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr17_-_18430160 | 0.00 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr15_+_58430368 | 0.00 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr3_+_186353756 | 0.00 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr11_-_117747434 | 0.00 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of VAX2_RHOXF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.8 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0044793 | negative regulation by host of viral process(GO:0044793) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:1904879 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.8 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |