Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
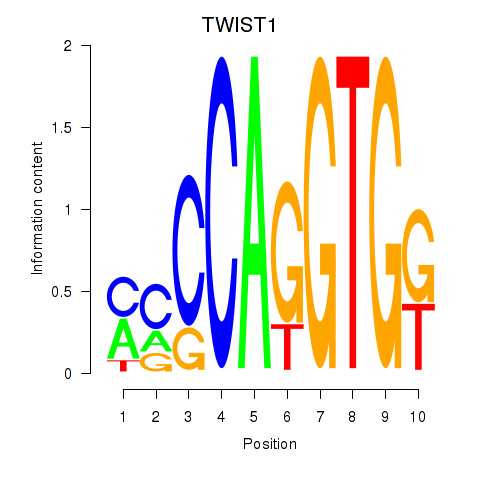
Results for TWIST1_SNAI1
Z-value: 0.42
Transcription factors associated with TWIST1_SNAI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TWIST1
|
ENSG00000122691.8 | twist family bHLH transcription factor 1 |
|
SNAI1
|
ENSG00000124216.3 | snail family transcriptional repressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TWIST1 | hg19_v2_chr7_-_19157248_19157295 | -0.78 | 7.0e-02 | Click! |
| SNAI1 | hg19_v2_chr20_+_48599506_48599536 | 0.43 | 4.0e-01 | Click! |
Activity profile of TWIST1_SNAI1 motif
Sorted Z-values of TWIST1_SNAI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_1850697 | 0.31 |
ENST00000378598.4
ENST00000416272.1 ENST00000310991.3 |
TMEM52
|
transmembrane protein 52 |
| chr7_-_1595756 | 0.21 |
ENST00000441933.1
|
TMEM184A
|
transmembrane protein 184A |
| chr17_+_39421591 | 0.20 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr17_-_42452063 | 0.18 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr19_-_7939319 | 0.17 |
ENST00000539422.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr17_+_39411636 | 0.16 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chr19_-_56109119 | 0.16 |
ENST00000587678.1
|
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr4_-_11430221 | 0.15 |
ENST00000514690.1
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr5_+_150406527 | 0.15 |
ENST00000520059.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr15_+_91416092 | 0.14 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr11_-_615570 | 0.14 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr12_+_8849773 | 0.14 |
ENST00000541044.1
|
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr11_-_118868682 | 0.14 |
ENST00000526453.1
|
RP11-110I1.12
|
RP11-110I1.12 |
| chr2_-_241500168 | 0.14 |
ENST00000443318.1
ENST00000411765.1 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr17_-_72968837 | 0.13 |
ENST00000581676.1
|
HID1
|
HID1 domain containing |
| chr16_-_30393752 | 0.12 |
ENST00000566517.1
ENST00000605106.1 |
SEPT1
SEPT1
|
septin 1 Uncharacterized protein |
| chr20_-_62493217 | 0.12 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135 |
| chr19_-_5567842 | 0.12 |
ENST00000587632.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr6_-_42110342 | 0.12 |
ENST00000356542.5
|
C6orf132
|
chromosome 6 open reading frame 132 |
| chr2_+_128177458 | 0.12 |
ENST00000409048.1
ENST00000422777.3 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr22_-_39096981 | 0.12 |
ENST00000427389.1
|
JOSD1
|
Josephin domain containing 1 |
| chr22_-_21905120 | 0.12 |
ENST00000331505.5
|
RIMBP3C
|
RIMS binding protein 3C |
| chr11_+_117049910 | 0.11 |
ENST00000431081.2
ENST00000524842.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr20_+_2795626 | 0.11 |
ENST00000603872.1
ENST00000380589.4 |
C20orf141
|
chromosome 20 open reading frame 141 |
| chr11_+_62379194 | 0.11 |
ENST00000525801.1
ENST00000534093.1 |
ROM1
|
retinal outer segment membrane protein 1 |
| chr6_+_159290917 | 0.11 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr19_+_10217364 | 0.11 |
ENST00000430370.1
|
PPAN
|
peter pan homolog (Drosophila) |
| chr11_+_826136 | 0.11 |
ENST00000528315.1
ENST00000533803.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr13_-_101240985 | 0.11 |
ENST00000471912.1
|
GGACT
|
gamma-glutamylamine cyclotransferase |
| chr20_+_48599506 | 0.10 |
ENST00000244050.2
|
SNAI1
|
snail family zinc finger 1 |
| chr12_+_112451222 | 0.10 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr7_-_1595871 | 0.10 |
ENST00000319010.5
|
TMEM184A
|
transmembrane protein 184A |
| chr7_-_127032114 | 0.10 |
ENST00000436992.1
|
ZNF800
|
zinc finger protein 800 |
| chr2_+_121493717 | 0.10 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr16_-_30394143 | 0.10 |
ENST00000321367.3
ENST00000571393.1 |
SEPT1
|
septin 1 |
| chr10_+_116853091 | 0.10 |
ENST00000526946.1
|
ATRNL1
|
attractin-like 1 |
| chr1_-_95285652 | 0.10 |
ENST00000442418.1
|
LINC01057
|
long intergenic non-protein coding RNA 1057 |
| chr17_-_71258019 | 0.10 |
ENST00000344935.4
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like |
| chr12_-_12714025 | 0.10 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr3_-_93747425 | 0.10 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr19_-_17932314 | 0.10 |
ENST00000598577.1
ENST00000317306.7 ENST00000379695.5 |
INSL3
|
insulin-like 3 (Leydig cell) |
| chr11_+_45168182 | 0.09 |
ENST00000526442.1
|
PRDM11
|
PR domain containing 11 |
| chrX_-_128782722 | 0.09 |
ENST00000427399.1
|
APLN
|
apelin |
| chr1_-_22215192 | 0.09 |
ENST00000374673.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr14_-_69263043 | 0.09 |
ENST00000408913.2
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr22_+_27068766 | 0.09 |
ENST00000435162.1
ENST00000437071.1 ENST00000440816.1 ENST00000421253.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr1_+_8378140 | 0.09 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr14_-_94789663 | 0.09 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr17_-_40835076 | 0.09 |
ENST00000591765.1
|
CCR10
|
chemokine (C-C motif) receptor 10 |
| chr19_+_17638059 | 0.09 |
ENST00000599164.1
ENST00000449408.2 ENST00000600871.1 ENST00000599124.1 |
FAM129C
|
family with sequence similarity 129, member C |
| chr17_-_70053866 | 0.09 |
ENST00000540802.1
|
RP11-84E24.2
|
RP11-84E24.2 |
| chr11_-_17410629 | 0.08 |
ENST00000526912.1
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11 |
| chr21_-_47352477 | 0.08 |
ENST00000593412.1
|
PRED62
|
Uncharacterized protein |
| chr7_+_76054224 | 0.08 |
ENST00000394857.3
|
ZP3
|
zona pellucida glycoprotein 3 (sperm receptor) |
| chr4_+_89299885 | 0.08 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr11_+_74870818 | 0.08 |
ENST00000525845.1
ENST00000534186.1 ENST00000428359.2 |
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr8_+_145065521 | 0.08 |
ENST00000534791.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr19_+_50433476 | 0.08 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr11_-_2924720 | 0.08 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr1_+_54359854 | 0.08 |
ENST00000361921.3
ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1
|
deiodinase, iodothyronine, type I |
| chr12_-_110318130 | 0.08 |
ENST00000540772.1
|
GLTP
|
glycolipid transfer protein |
| chr8_+_145065705 | 0.07 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr3_-_48481518 | 0.07 |
ENST00000412398.2
ENST00000395696.1 |
CCDC51
|
coiled-coil domain containing 51 |
| chr19_-_53636125 | 0.07 |
ENST00000601493.1
ENST00000599261.1 ENST00000597503.1 ENST00000500065.4 ENST00000243643.4 ENST00000594011.1 ENST00000455735.2 ENST00000595193.1 ENST00000448501.1 ENST00000421033.1 ENST00000440291.1 ENST00000595813.1 ENST00000600574.1 ENST00000596051.1 ENST00000601110.1 |
ZNF415
|
zinc finger protein 415 |
| chr12_-_95942563 | 0.07 |
ENST00000549639.1
ENST00000551837.1 |
USP44
|
ubiquitin specific peptidase 44 |
| chr11_-_65626753 | 0.07 |
ENST00000526975.1
ENST00000531413.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr17_+_77030267 | 0.07 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr6_-_44225231 | 0.07 |
ENST00000538577.1
ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr19_+_36359341 | 0.07 |
ENST00000221891.4
|
APLP1
|
amyloid beta (A4) precursor-like protein 1 |
| chr6_+_75994755 | 0.07 |
ENST00000607799.1
|
RP1-234P15.4
|
RP1-234P15.4 |
| chr11_-_118927561 | 0.07 |
ENST00000530473.1
|
HYOU1
|
hypoxia up-regulated 1 |
| chr22_+_24820341 | 0.07 |
ENST00000464977.1
ENST00000444262.2 |
ADORA2A
|
adenosine A2a receptor |
| chr16_+_4838393 | 0.07 |
ENST00000589721.1
|
SMIM22
|
small integral membrane protein 22 |
| chr15_+_42565393 | 0.07 |
ENST00000561871.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr7_-_120497178 | 0.07 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr19_-_6375860 | 0.07 |
ENST00000245810.1
|
PSPN
|
persephin |
| chr6_-_31138439 | 0.07 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr16_+_718147 | 0.07 |
ENST00000561929.1
|
RHOT2
|
ras homolog family member T2 |
| chr2_+_128177253 | 0.07 |
ENST00000427769.1
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr12_+_82752283 | 0.07 |
ENST00000548200.1
|
METTL25
|
methyltransferase like 25 |
| chr16_+_2106134 | 0.07 |
ENST00000467949.1
|
TSC2
|
tuberous sclerosis 2 |
| chr5_+_132083106 | 0.07 |
ENST00000378731.1
|
CCNI2
|
cyclin I family, member 2 |
| chr3_-_48481434 | 0.07 |
ENST00000395694.2
ENST00000447018.1 ENST00000442740.1 |
CCDC51
|
coiled-coil domain containing 51 |
| chr11_-_57102947 | 0.07 |
ENST00000526696.1
|
SSRP1
|
structure specific recognition protein 1 |
| chr7_-_127672146 | 0.07 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr11_+_94439591 | 0.07 |
ENST00000299004.9
|
AMOTL1
|
angiomotin like 1 |
| chr17_-_79166176 | 0.07 |
ENST00000571292.1
|
AZI1
|
5-azacytidine induced 1 |
| chr7_-_1595107 | 0.07 |
ENST00000414730.1
|
TMEM184A
|
transmembrane protein 184A |
| chr22_+_19467261 | 0.07 |
ENST00000455750.1
ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45
|
cell division cycle 45 |
| chr8_+_22429205 | 0.06 |
ENST00000520207.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr2_+_172864490 | 0.06 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr10_+_47658234 | 0.06 |
ENST00000447511.2
ENST00000537271.1 |
ANTXRL
|
anthrax toxin receptor-like |
| chr4_+_8201091 | 0.06 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr17_-_33469299 | 0.06 |
ENST00000586869.1
ENST00000360831.5 ENST00000442241.4 |
NLE1
|
notchless homolog 1 (Drosophila) |
| chr17_+_7533439 | 0.06 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr10_+_99258625 | 0.06 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr5_-_134375386 | 0.06 |
ENST00000511256.1
|
CTC-276P9.1
|
CTC-276P9.1 |
| chr9_+_108424738 | 0.06 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr14_-_23652849 | 0.06 |
ENST00000316902.7
ENST00000469263.1 ENST00000525062.1 ENST00000524758.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr8_+_58055238 | 0.06 |
ENST00000519314.1
ENST00000519241.1 |
RP11-513O17.2
|
RP11-513O17.2 |
| chr19_-_40730820 | 0.06 |
ENST00000513948.1
|
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr12_+_116955659 | 0.06 |
ENST00000552992.1
|
RP11-148B3.1
|
RP11-148B3.1 |
| chr1_+_228337553 | 0.06 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr21_+_37507210 | 0.06 |
ENST00000290354.5
|
CBR3
|
carbonyl reductase 3 |
| chr7_-_135433460 | 0.06 |
ENST00000415751.1
|
FAM180A
|
family with sequence similarity 180, member A |
| chr19_-_55895966 | 0.06 |
ENST00000444469.3
|
TMEM238
|
transmembrane protein 238 |
| chr12_+_38710555 | 0.06 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chr17_+_78193443 | 0.06 |
ENST00000577155.1
|
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr5_-_102455801 | 0.06 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr1_-_169680745 | 0.06 |
ENST00000236147.4
|
SELL
|
selectin L |
| chr19_+_18794470 | 0.06 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr18_+_47087390 | 0.06 |
ENST00000583083.1
|
LIPG
|
lipase, endothelial |
| chr17_+_42015654 | 0.06 |
ENST00000565120.1
|
RP11-527L4.2
|
Uncharacterized protein |
| chr16_-_89268070 | 0.06 |
ENST00000562855.2
|
SLC22A31
|
solute carrier family 22, member 31 |
| chr3_+_49059038 | 0.06 |
ENST00000451378.2
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr13_+_111972980 | 0.05 |
ENST00000283547.1
|
TEX29
|
testis expressed 29 |
| chr2_-_74776586 | 0.05 |
ENST00000420535.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr6_+_63921399 | 0.05 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr3_+_44596679 | 0.05 |
ENST00000426540.1
ENST00000431636.1 ENST00000341840.3 ENST00000273320.3 |
ZKSCAN7
|
zinc finger with KRAB and SCAN domains 7 |
| chr7_+_1084206 | 0.05 |
ENST00000444847.1
|
GPR146
|
G protein-coupled receptor 146 |
| chr19_-_5567996 | 0.05 |
ENST00000448587.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr17_+_78194261 | 0.05 |
ENST00000572725.1
|
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr3_-_197282821 | 0.05 |
ENST00000445160.2
ENST00000446746.1 ENST00000432819.1 ENST00000392379.1 ENST00000441275.1 ENST00000392378.2 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr19_-_1168936 | 0.05 |
ENST00000587655.1
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr15_-_78112553 | 0.05 |
ENST00000562933.1
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr19_+_35630926 | 0.05 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr2_-_74669009 | 0.05 |
ENST00000272430.5
|
RTKN
|
rhotekin |
| chr10_+_73156664 | 0.05 |
ENST00000398809.4
ENST00000398842.3 ENST00000461841.3 ENST00000299366.7 |
CDH23
|
cadherin-related 23 |
| chr9_+_92219919 | 0.05 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr14_-_37051798 | 0.05 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr4_+_89299994 | 0.05 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr1_+_92495528 | 0.05 |
ENST00000370383.4
|
EPHX4
|
epoxide hydrolase 4 |
| chr12_+_52626898 | 0.05 |
ENST00000331817.5
|
KRT7
|
keratin 7 |
| chr1_+_3773825 | 0.05 |
ENST00000378209.3
ENST00000338895.3 ENST00000378212.2 ENST00000341385.3 |
DFFB
|
DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) |
| chr21_-_43916296 | 0.05 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr19_+_50691437 | 0.05 |
ENST00000598205.1
|
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr19_-_33793430 | 0.05 |
ENST00000498907.2
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr22_+_39868786 | 0.05 |
ENST00000429402.1
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr6_-_3227877 | 0.05 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb |
| chr12_-_51611477 | 0.05 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr6_+_27215494 | 0.05 |
ENST00000230582.3
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr17_+_74075263 | 0.05 |
ENST00000334586.5
ENST00000392503.2 |
ZACN
|
zinc activated ligand-gated ion channel |
| chr11_-_119293903 | 0.05 |
ENST00000580275.1
|
THY1
|
Thy-1 cell surface antigen |
| chr1_-_154178803 | 0.05 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr12_+_110172572 | 0.05 |
ENST00000358906.3
|
FAM222A
|
family with sequence similarity 222, member A |
| chr19_-_50432711 | 0.05 |
ENST00000597723.1
ENST00000599788.1 ENST00000596217.1 ENST00000593652.1 ENST00000599567.1 ENST00000600935.1 ENST00000596011.1 ENST00000596022.1 ENST00000597295.1 |
NUP62
IL4I1
|
nucleoporin 62kDa interleukin 4 induced 1 |
| chr1_+_11714425 | 0.05 |
ENST00000251546.4
|
FBXO44
|
F-box protein 44 |
| chr11_-_118972575 | 0.05 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr8_+_22438009 | 0.05 |
ENST00000409417.1
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr1_-_32827682 | 0.05 |
ENST00000432622.1
|
FAM229A
|
family with sequence similarity 229, member A |
| chr17_+_75277492 | 0.05 |
ENST00000427177.1
ENST00000591198.1 |
SEPT9
|
septin 9 |
| chr16_+_718086 | 0.05 |
ENST00000315082.4
ENST00000563134.1 |
RHOT2
|
ras homolog family member T2 |
| chr12_-_110318263 | 0.05 |
ENST00000318348.4
|
GLTP
|
glycolipid transfer protein |
| chr3_+_42695176 | 0.05 |
ENST00000232974.6
ENST00000457842.3 |
ZBTB47
|
zinc finger and BTB domain containing 47 |
| chr11_-_65626797 | 0.05 |
ENST00000525451.2
|
CFL1
|
cofilin 1 (non-muscle) |
| chr20_+_49348109 | 0.04 |
ENST00000396039.1
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr11_+_117049854 | 0.04 |
ENST00000278951.7
|
SIDT2
|
SID1 transmembrane family, member 2 |
| chr7_-_2883928 | 0.04 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr5_-_132948216 | 0.04 |
ENST00000265342.7
|
FSTL4
|
follistatin-like 4 |
| chr1_-_201123546 | 0.04 |
ENST00000435310.1
ENST00000485839.2 ENST00000367330.1 |
TMEM9
|
transmembrane protein 9 |
| chr19_-_18391708 | 0.04 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chrX_-_153775426 | 0.04 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr6_-_31550192 | 0.04 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr4_-_40632140 | 0.04 |
ENST00000514782.1
|
RBM47
|
RNA binding motif protein 47 |
| chr6_+_11537910 | 0.04 |
ENST00000543875.1
|
TMEM170B
|
transmembrane protein 170B |
| chr22_+_41697520 | 0.04 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr12_+_122516626 | 0.04 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chrX_-_153059958 | 0.04 |
ENST00000370092.3
ENST00000217901.5 |
IDH3G
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr11_-_119993979 | 0.04 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr7_+_142982023 | 0.04 |
ENST00000359333.3
ENST00000409244.1 ENST00000409541.1 ENST00000410004.1 |
TMEM139
|
transmembrane protein 139 |
| chr1_+_14026671 | 0.04 |
ENST00000484063.2
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr11_+_65779283 | 0.04 |
ENST00000312134.2
|
CST6
|
cystatin E/M |
| chr19_-_11545920 | 0.04 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr22_+_22681656 | 0.04 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr19_+_49055332 | 0.04 |
ENST00000201586.2
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1 |
| chr3_-_52002194 | 0.04 |
ENST00000466412.1
|
PCBP4
|
poly(rC) binding protein 4 |
| chr2_+_191045656 | 0.04 |
ENST00000443551.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr9_+_138392483 | 0.04 |
ENST00000241600.5
|
MRPS2
|
mitochondrial ribosomal protein S2 |
| chr1_-_1149506 | 0.04 |
ENST00000379236.3
|
TNFRSF4
|
tumor necrosis factor receptor superfamily, member 4 |
| chr17_+_73539339 | 0.04 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr19_-_50432782 | 0.04 |
ENST00000413454.1
ENST00000596437.1 ENST00000341114.3 ENST00000595948.1 |
NUP62
IL4I1
|
nucleoporin 62kDa interleukin 4 induced 1 |
| chr9_+_139221880 | 0.04 |
ENST00000392945.3
ENST00000440944.1 |
GPSM1
|
G-protein signaling modulator 1 |
| chr16_-_4838255 | 0.04 |
ENST00000591624.1
ENST00000396693.5 |
SEPT12
|
septin 12 |
| chr19_+_39897943 | 0.04 |
ENST00000600033.1
|
ZFP36
|
ZFP36 ring finger protein |
| chr19_-_10047219 | 0.04 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr8_+_22436635 | 0.04 |
ENST00000452226.1
ENST00000397760.4 ENST00000339162.7 ENST00000397761.2 |
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr5_-_150467221 | 0.04 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr22_-_19974616 | 0.04 |
ENST00000344269.3
ENST00000401994.1 ENST00000406522.1 |
ARVCF
|
armadillo repeat gene deleted in velocardiofacial syndrome |
| chr22_-_39096661 | 0.04 |
ENST00000216039.5
|
JOSD1
|
Josephin domain containing 1 |
| chr8_+_67687773 | 0.04 |
ENST00000518388.1
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr14_-_81902791 | 0.04 |
ENST00000557055.1
|
STON2
|
stonin 2 |
| chr19_+_39989535 | 0.04 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr11_+_66115304 | 0.04 |
ENST00000531602.1
|
RP11-867G23.8
|
Uncharacterized protein |
| chr9_-_140142222 | 0.04 |
ENST00000344774.4
ENST00000388932.2 |
FAM166A
|
family with sequence similarity 166, member A |
| chr20_-_3767324 | 0.04 |
ENST00000379751.4
|
CENPB
|
centromere protein B, 80kDa |
| chr10_-_69991865 | 0.04 |
ENST00000373673.3
|
ATOH7
|
atonal homolog 7 (Drosophila) |
| chr8_+_22436248 | 0.04 |
ENST00000308354.7
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr1_+_10509971 | 0.04 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr17_+_29815013 | 0.04 |
ENST00000394744.2
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II) |
| chr2_-_86564740 | 0.04 |
ENST00000540790.1
ENST00000428491.1 |
REEP1
|
receptor accessory protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TWIST1_SNAI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.1 | GO:0032904 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.2 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.2 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.1 | GO:1903381 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.0 | 0.1 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.1 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.2 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.0 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0051511 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.0 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.0 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.0 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.0 | 0.1 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.0 | GO:0001923 | B-1 B cell differentiation(GO:0001923) B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.0 | GO:0060929 | Purkinje myocyte differentiation(GO:0003168) cardiac pacemaker cell fate commitment(GO:0060927) atrioventricular node cell fate commitment(GO:0060929) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.0 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.0 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |