Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
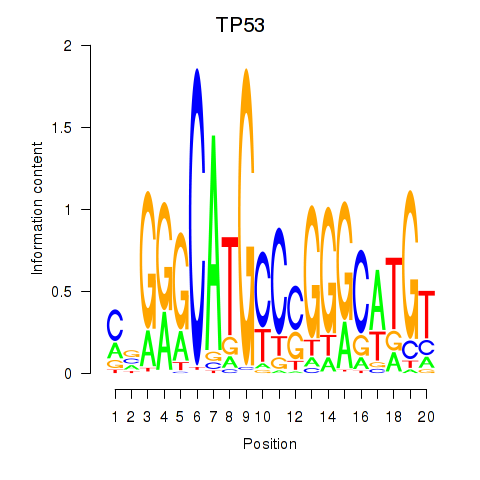
Results for TP53
Z-value: 1.14
Transcription factors associated with TP53
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP53
|
ENSG00000141510.11 | tumor protein p53 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP53 | hg19_v2_chr17_-_7590745_7590856 | 0.65 | 1.6e-01 | Click! |
Activity profile of TP53 motif
Sorted Z-values of TP53 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_139847347 | 0.97 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr16_-_75467274 | 0.92 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1 |
| chr10_-_105212141 | 0.82 |
ENST00000369788.3
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr10_-_105212059 | 0.70 |
ENST00000260743.5
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr19_+_13988061 | 0.60 |
ENST00000339133.5
ENST00000397555.2 |
NANOS3
|
nanos homolog 3 (Drosophila) |
| chr16_-_4303767 | 0.51 |
ENST00000573268.1
ENST00000573042.1 |
RP11-95P2.1
|
RP11-95P2.1 |
| chr9_-_140115775 | 0.51 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr19_+_827823 | 0.37 |
ENST00000233997.2
|
AZU1
|
azurocidin 1 |
| chr15_+_100347228 | 0.37 |
ENST00000559714.1
ENST00000560059.1 |
CTD-2054N24.2
|
Uncharacterized protein |
| chr19_-_5567842 | 0.33 |
ENST00000587632.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr12_-_55042140 | 0.33 |
ENST00000293371.6
ENST00000456047.2 |
DCD
|
dermcidin |
| chr13_-_48877795 | 0.32 |
ENST00000436963.1
ENST00000433480.2 |
LINC00441
|
long intergenic non-protein coding RNA 441 |
| chr17_-_74528128 | 0.32 |
ENST00000590175.1
|
CYGB
|
cytoglobin |
| chr1_+_116519223 | 0.32 |
ENST00000369502.1
|
SLC22A15
|
solute carrier family 22, member 15 |
| chr1_-_10532531 | 0.31 |
ENST00000377036.2
ENST00000377038.3 |
DFFA
|
DNA fragmentation factor, 45kDa, alpha polypeptide |
| chr1_+_204494618 | 0.30 |
ENST00000367180.1
ENST00000391947.2 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr22_+_29168652 | 0.30 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr2_-_75788424 | 0.30 |
ENST00000410071.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr17_+_14204389 | 0.28 |
ENST00000360954.2
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
| chr9_+_130186653 | 0.27 |
ENST00000342483.5
ENST00000543471.1 |
ZNF79
|
zinc finger protein 79 |
| chr5_+_178487354 | 0.27 |
ENST00000315475.6
|
ZNF354C
|
zinc finger protein 354C |
| chr1_-_169396666 | 0.26 |
ENST00000456107.1
ENST00000367805.3 |
CCDC181
|
coiled-coil domain containing 181 |
| chr17_+_76210367 | 0.26 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr22_-_38484922 | 0.25 |
ENST00000428572.1
|
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr1_+_10057274 | 0.24 |
ENST00000294435.7
|
RBP7
|
retinol binding protein 7, cellular |
| chr9_-_115774453 | 0.24 |
ENST00000427548.1
|
ZNF883
|
zinc finger protein 883 |
| chr19_+_51293672 | 0.23 |
ENST00000270593.1
ENST00000270594.3 |
ACPT
|
acid phosphatase, testicular |
| chr15_-_102285913 | 0.23 |
ENST00000558592.1
|
RP11-89K11.1
|
Uncharacterized protein |
| chr2_-_233286046 | 0.22 |
ENST00000437095.1
|
AC068134.6
|
AC068134.6 |
| chr1_-_156051789 | 0.22 |
ENST00000532414.2
|
MEX3A
|
mex-3 RNA binding family member A |
| chr19_-_28284793 | 0.22 |
ENST00000590523.1
|
LINC00662
|
long intergenic non-protein coding RNA 662 |
| chr20_-_23669590 | 0.21 |
ENST00000217423.3
|
CST4
|
cystatin S |
| chr19_-_38714847 | 0.21 |
ENST00000420980.2
ENST00000355526.4 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr20_+_62289640 | 0.21 |
ENST00000508582.2
ENST00000360203.5 ENST00000356810.4 |
RTEL1
|
regulator of telomere elongation helicase 1 |
| chr2_-_157198860 | 0.20 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr4_+_186347388 | 0.20 |
ENST00000511138.1
ENST00000511581.1 |
C4orf47
|
chromosome 4 open reading frame 47 |
| chr16_+_77756399 | 0.20 |
ENST00000564085.1
ENST00000268533.5 ENST00000568787.1 ENST00000437314.3 ENST00000563839.1 |
NUDT7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr9_-_138853156 | 0.20 |
ENST00000371756.3
|
UBAC1
|
UBA domain containing 1 |
| chr22_-_28392227 | 0.19 |
ENST00000431039.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr5_-_131826457 | 0.19 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr12_+_44229846 | 0.19 |
ENST00000551577.1
ENST00000266534.3 |
TMEM117
|
transmembrane protein 117 |
| chr11_-_73309228 | 0.18 |
ENST00000356467.4
ENST00000064778.4 |
FAM168A
|
family with sequence similarity 168, member A |
| chr19_+_18485509 | 0.18 |
ENST00000597765.1
|
GDF15
|
growth differentiation factor 15 |
| chr7_+_72742178 | 0.18 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr2_+_10281509 | 0.18 |
ENST00000381786.3
|
C2orf48
|
chromosome 2 open reading frame 48 |
| chr19_-_44258770 | 0.18 |
ENST00000601925.1
ENST00000602222.1 ENST00000599804.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr19_+_39687596 | 0.18 |
ENST00000339852.4
|
NCCRP1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr17_+_73455788 | 0.17 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr19_+_45174724 | 0.17 |
ENST00000358777.4
|
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr2_-_75788038 | 0.17 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr19_+_57874835 | 0.17 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr16_+_2079637 | 0.17 |
ENST00000561844.1
|
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr19_+_39759154 | 0.17 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr10_+_134000404 | 0.17 |
ENST00000338492.4
ENST00000368629.1 |
DPYSL4
|
dihydropyrimidinase-like 4 |
| chr19_+_926000 | 0.17 |
ENST00000263620.3
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr7_-_74221288 | 0.17 |
ENST00000451013.2
|
GTF2IRD2
|
GTF2I repeat domain containing 2 |
| chr19_+_3136115 | 0.16 |
ENST00000262958.3
|
GNA15
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
| chr19_+_28284803 | 0.16 |
ENST00000586220.1
ENST00000588784.1 ENST00000591549.1 ENST00000585827.1 ENST00000588636.1 ENST00000587188.1 |
CTC-459F4.3
|
CTC-459F4.3 |
| chr3_-_52479043 | 0.15 |
ENST00000231721.2
ENST00000475739.1 |
SEMA3G
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| chrX_+_192989 | 0.15 |
ENST00000399012.1
ENST00000430923.2 |
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr1_-_201438282 | 0.15 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr20_-_23807358 | 0.15 |
ENST00000304725.2
|
CST2
|
cystatin SA |
| chr4_-_40516560 | 0.14 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr22_+_40766582 | 0.14 |
ENST00000457767.1
ENST00000248929.9 ENST00000454798.2 |
SGSM3
|
small G protein signaling modulator 3 |
| chr22_+_19705928 | 0.14 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chr19_-_44258733 | 0.14 |
ENST00000597586.1
ENST00000596714.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr11_+_289110 | 0.14 |
ENST00000409548.2
|
ATHL1
|
ATH1, acid trehalase-like 1 (yeast) |
| chr1_-_45805752 | 0.14 |
ENST00000354383.6
ENST00000355498.2 ENST00000372100.5 ENST00000531105.1 |
MUTYH
|
mutY homolog |
| chr19_-_45735138 | 0.14 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr12_+_69202975 | 0.13 |
ENST00000544561.1
ENST00000393410.1 ENST00000299252.4 ENST00000360430.2 ENST00000517852.1 ENST00000545204.1 ENST00000393413.3 ENST00000350057.5 ENST00000348801.2 ENST00000478070.1 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr1_-_38412683 | 0.13 |
ENST00000373024.3
ENST00000373023.2 |
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa |
| chr4_-_122686261 | 0.13 |
ENST00000337677.5
|
TMEM155
|
transmembrane protein 155 |
| chr22_+_22901750 | 0.13 |
ENST00000407120.1
|
LL22NC03-63E9.3
|
Uncharacterized protein |
| chr3_+_44379944 | 0.13 |
ENST00000396078.3
ENST00000342649.4 |
TCAIM
|
T cell activation inhibitor, mitochondrial |
| chr14_+_36295638 | 0.13 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr7_-_100493482 | 0.13 |
ENST00000411582.1
ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr15_+_89164560 | 0.12 |
ENST00000379231.3
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr8_-_12612962 | 0.12 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr19_+_45174994 | 0.12 |
ENST00000403660.3
|
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr3_-_100120223 | 0.12 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr14_-_103989033 | 0.12 |
ENST00000553878.1
ENST00000557530.1 |
CKB
|
creatine kinase, brain |
| chr1_+_116519112 | 0.12 |
ENST00000369503.4
|
SLC22A15
|
solute carrier family 22, member 15 |
| chr6_+_33378517 | 0.12 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1 |
| chr3_-_46037299 | 0.12 |
ENST00000296137.2
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr14_-_91282821 | 0.12 |
ENST00000553948.1
|
TTC7B
|
tetratricopeptide repeat domain 7B |
| chr2_-_242556900 | 0.12 |
ENST00000402545.1
ENST00000402136.1 |
THAP4
|
THAP domain containing 4 |
| chr1_-_1356628 | 0.12 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr11_+_119038897 | 0.12 |
ENST00000454811.1
ENST00000449394.1 |
NLRX1
|
NLR family member X1 |
| chr16_+_15744078 | 0.11 |
ENST00000396354.1
ENST00000570727.1 |
NDE1
|
nudE neurodevelopment protein 1 |
| chr12_-_126467906 | 0.11 |
ENST00000507313.1
ENST00000545784.1 |
LINC00939
|
long intergenic non-protein coding RNA 939 |
| chr18_-_28622774 | 0.11 |
ENST00000434452.1
|
DSC3
|
desmocollin 3 |
| chr1_+_25598872 | 0.11 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr11_+_64879317 | 0.11 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr10_-_47173994 | 0.11 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr1_-_1356719 | 0.11 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr1_+_211432700 | 0.11 |
ENST00000452621.2
|
RCOR3
|
REST corepressor 3 |
| chr18_-_28622699 | 0.11 |
ENST00000360428.4
|
DSC3
|
desmocollin 3 |
| chr12_+_53894672 | 0.11 |
ENST00000266987.2
ENST00000456234.2 |
TARBP2
|
TAR (HIV-1) RNA binding protein 2 |
| chr18_+_44497455 | 0.11 |
ENST00000592005.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr1_+_2477831 | 0.11 |
ENST00000606645.1
|
RP3-395M20.12
|
RP3-395M20.12 |
| chr10_+_64564469 | 0.10 |
ENST00000373783.1
|
ADO
|
2-aminoethanethiol (cysteamine) dioxygenase |
| chr9_-_135996537 | 0.10 |
ENST00000372050.3
ENST00000372047.3 |
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr9_+_139846708 | 0.10 |
ENST00000371633.3
|
LCN12
|
lipocalin 12 |
| chr11_+_64002292 | 0.10 |
ENST00000426086.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr19_-_51142540 | 0.10 |
ENST00000598997.1
|
SYT3
|
synaptotagmin III |
| chr17_+_74372662 | 0.10 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr19_-_55690758 | 0.10 |
ENST00000590851.1
|
SYT5
|
synaptotagmin V |
| chr16_+_2079501 | 0.10 |
ENST00000563587.1
|
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr2_+_11295624 | 0.10 |
ENST00000402361.1
ENST00000428481.1 |
PQLC3
|
PQ loop repeat containing 3 |
| chr6_-_165723088 | 0.10 |
ENST00000230301.8
|
C6orf118
|
chromosome 6 open reading frame 118 |
| chr1_-_2458026 | 0.10 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr3_+_122785895 | 0.10 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr12_-_6451014 | 0.09 |
ENST00000366159.4
ENST00000539372.1 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chrX_+_37545012 | 0.09 |
ENST00000378616.3
|
XK
|
X-linked Kx blood group (McLeod syndrome) |
| chr6_+_52285131 | 0.09 |
ENST00000433625.2
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr6_+_33378738 | 0.09 |
ENST00000374512.3
ENST00000374516.3 |
PHF1
|
PHD finger protein 1 |
| chr1_-_45806053 | 0.09 |
ENST00000412971.1
ENST00000372098.3 ENST00000372110.3 ENST00000529984.1 ENST00000528332.2 ENST00000372115.3 ENST00000450313.1 |
MUTYH
|
mutY homolog |
| chr12_-_6451235 | 0.09 |
ENST00000440083.2
ENST00000162749.2 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr2_+_47168630 | 0.09 |
ENST00000263737.6
|
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr4_+_52709229 | 0.09 |
ENST00000334635.5
ENST00000381441.3 ENST00000381437.4 |
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 |
| chr3_+_49977894 | 0.09 |
ENST00000433811.1
|
RBM6
|
RNA binding motif protein 6 |
| chr19_-_44259053 | 0.09 |
ENST00000601170.1
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr1_-_26197744 | 0.09 |
ENST00000374296.3
|
PAQR7
|
progestin and adipoQ receptor family member VII |
| chr2_-_75788428 | 0.09 |
ENST00000432649.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr3_+_49977490 | 0.09 |
ENST00000539992.1
|
RBM6
|
RNA binding motif protein 6 |
| chr17_+_12859080 | 0.09 |
ENST00000583608.1
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr15_-_30685563 | 0.09 |
ENST00000401522.3
|
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr4_-_10117949 | 0.09 |
ENST00000508079.1
|
WDR1
|
WD repeat domain 1 |
| chr3_-_44519131 | 0.08 |
ENST00000425708.2
ENST00000396077.2 |
ZNF445
|
zinc finger protein 445 |
| chr2_+_197504278 | 0.08 |
ENST00000272831.7
ENST00000389175.4 ENST00000472405.2 ENST00000423093.2 |
CCDC150
|
coiled-coil domain containing 150 |
| chr2_+_47168313 | 0.08 |
ENST00000319190.5
ENST00000394850.2 ENST00000536057.1 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr16_-_29874211 | 0.08 |
ENST00000563415.1
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr19_-_44259136 | 0.08 |
ENST00000270066.6
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr19_-_40931891 | 0.08 |
ENST00000357949.4
|
SERTAD1
|
SERTA domain containing 1 |
| chr16_+_57653625 | 0.08 |
ENST00000567553.1
ENST00000565314.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr1_-_6453399 | 0.08 |
ENST00000608083.1
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr3_+_149530469 | 0.08 |
ENST00000392894.3
ENST00000344229.3 |
RNF13
|
ring finger protein 13 |
| chr13_-_102068706 | 0.08 |
ENST00000251127.6
|
NALCN
|
sodium leak channel, non-selective |
| chr17_-_7387524 | 0.08 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr11_+_65222698 | 0.08 |
ENST00000309775.7
|
AP000769.1
|
Uncharacterized protein |
| chr7_-_100239132 | 0.08 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr12_+_54519842 | 0.08 |
ENST00000508564.1
|
RP11-834C11.4
|
RP11-834C11.4 |
| chr3_+_14166440 | 0.07 |
ENST00000306077.4
|
TMEM43
|
transmembrane protein 43 |
| chr11_-_82997013 | 0.07 |
ENST00000529073.1
ENST00000529611.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr9_+_139877445 | 0.07 |
ENST00000408973.2
|
LCNL1
|
lipocalin-like 1 |
| chr6_+_117803797 | 0.07 |
ENST00000296955.8
ENST00000368503.4 ENST00000338728.5 |
DCBLD1
|
discoidin, CUB and LCCL domain containing 1 |
| chr16_-_68000717 | 0.07 |
ENST00000541864.2
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr17_-_5095126 | 0.07 |
ENST00000576772.1
ENST00000575779.1 |
ZNF594
|
zinc finger protein 594 |
| chr17_-_61523622 | 0.07 |
ENST00000448884.2
ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561
|
cytochrome b561 |
| chr3_-_196045127 | 0.07 |
ENST00000325318.5
|
TCTEX1D2
|
Tctex1 domain containing 2 |
| chr9_+_132094579 | 0.07 |
ENST00000427109.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr16_+_57653854 | 0.07 |
ENST00000568908.1
ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr3_+_49977440 | 0.07 |
ENST00000442092.1
ENST00000266022.4 ENST00000443081.1 |
RBM6
|
RNA binding motif protein 6 |
| chr20_-_23618582 | 0.07 |
ENST00000398411.1
ENST00000376925.3 |
CST3
|
cystatin C |
| chr20_-_47804894 | 0.07 |
ENST00000371828.3
ENST00000371856.2 ENST00000360426.4 ENST00000347458.5 ENST00000340954.7 ENST00000371802.1 ENST00000371792.1 ENST00000437404.2 |
STAU1
|
staufen double-stranded RNA binding protein 1 |
| chr1_+_28206150 | 0.07 |
ENST00000456990.1
|
THEMIS2
|
thymocyte selection associated family member 2 |
| chr12_-_6451186 | 0.07 |
ENST00000540022.1
ENST00000536194.1 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr1_+_13910194 | 0.07 |
ENST00000376057.4
ENST00000510906.1 |
PDPN
|
podoplanin |
| chr5_-_95297534 | 0.07 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr12_+_53895052 | 0.07 |
ENST00000552857.1
|
TARBP2
|
TAR (HIV-1) RNA binding protein 2 |
| chr3_-_184079382 | 0.07 |
ENST00000344937.7
ENST00000423355.2 ENST00000434054.2 ENST00000457512.1 ENST00000265593.4 |
CLCN2
|
chloride channel, voltage-sensitive 2 |
| chr17_+_73089382 | 0.06 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr6_+_36098262 | 0.06 |
ENST00000373761.6
ENST00000373766.5 |
MAPK13
|
mitogen-activated protein kinase 13 |
| chrX_-_54209640 | 0.06 |
ENST00000375180.2
ENST00000328235.4 ENST00000477084.1 |
FAM120C
|
family with sequence similarity 120C |
| chr7_+_72742162 | 0.06 |
ENST00000431982.2
|
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr17_-_61523535 | 0.06 |
ENST00000584031.1
ENST00000392976.1 |
CYB561
|
cytochrome b561 |
| chr16_-_15149828 | 0.06 |
ENST00000566419.1
ENST00000568320.1 |
NTAN1
|
N-terminal asparagine amidase |
| chr6_-_101329157 | 0.06 |
ENST00000369143.2
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr1_+_15764931 | 0.06 |
ENST00000375949.4
ENST00000375943.2 |
CTRC
|
chymotrypsin C (caldecrin) |
| chr13_+_76334567 | 0.06 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr19_-_2456922 | 0.06 |
ENST00000582871.1
ENST00000325327.3 |
LMNB2
|
lamin B2 |
| chr1_-_202936394 | 0.06 |
ENST00000367249.4
|
CYB5R1
|
cytochrome b5 reductase 1 |
| chr16_+_57406368 | 0.06 |
ENST00000006053.6
ENST00000563383.1 |
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr19_-_17014408 | 0.06 |
ENST00000594249.1
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr16_-_29874462 | 0.06 |
ENST00000566113.1
ENST00000569956.1 ENST00000570016.1 |
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr5_-_168006324 | 0.06 |
ENST00000522176.1
|
PANK3
|
pantothenate kinase 3 |
| chr19_+_49458107 | 0.06 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr5_+_121647386 | 0.06 |
ENST00000542191.1
ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr4_-_129209944 | 0.06 |
ENST00000520121.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chrX_-_153583257 | 0.06 |
ENST00000438732.1
|
FLNA
|
filamin A, alpha |
| chr16_+_57653989 | 0.06 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr17_-_37309480 | 0.06 |
ENST00000539608.1
|
PLXDC1
|
plexin domain containing 1 |
| chr12_+_124086632 | 0.06 |
ENST00000238146.4
ENST00000538744.1 |
DDX55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr19_+_13229126 | 0.06 |
ENST00000292431.4
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr5_+_162864575 | 0.06 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr22_+_21336267 | 0.06 |
ENST00000215739.8
|
LZTR1
|
leucine-zipper-like transcription regulator 1 |
| chr17_-_37308824 | 0.05 |
ENST00000415163.1
ENST00000441877.1 ENST00000444911.2 |
PLXDC1
|
plexin domain containing 1 |
| chr7_+_23145884 | 0.05 |
ENST00000409689.1
ENST00000410047.1 |
KLHL7
|
kelch-like family member 7 |
| chr1_+_44115814 | 0.05 |
ENST00000372396.3
|
KDM4A
|
lysine (K)-specific demethylase 4A |
| chr19_+_18492973 | 0.05 |
ENST00000595973.2
|
GDF15
|
growth differentiation factor 15 |
| chr8_+_95732095 | 0.05 |
ENST00000414645.2
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chr9_-_95527079 | 0.05 |
ENST00000356884.6
ENST00000375512.3 |
BICD2
|
bicaudal D homolog 2 (Drosophila) |
| chr19_-_5567996 | 0.05 |
ENST00000448587.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr19_-_6591113 | 0.05 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr17_-_45918539 | 0.05 |
ENST00000584123.1
ENST00000578323.1 ENST00000407215.3 ENST00000290216.9 |
SCRN2
|
secernin 2 |
| chr19_-_47735918 | 0.05 |
ENST00000449228.1
ENST00000300880.7 ENST00000341983.4 |
BBC3
|
BCL2 binding component 3 |
| chr16_-_31106211 | 0.05 |
ENST00000532364.1
ENST00000529564.1 ENST00000319788.7 ENST00000354895.4 ENST00000394975.2 |
RP11-196G11.1
VKORC1
|
Uncharacterized protein vitamin K epoxide reductase complex, subunit 1 |
| chr19_+_3585471 | 0.05 |
ENST00000322315.5
|
GIPC3
|
GIPC PDZ domain containing family, member 3 |
| chr14_-_50999190 | 0.05 |
ENST00000557390.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr16_+_30207122 | 0.05 |
ENST00000395137.2
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP53
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.8 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.2 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.1 | 0.2 | GO:0045590 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.2 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.2 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.3 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.3 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.1 | GO:0060491 | regulation of cell projection assembly(GO:0060491) regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.2 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.6 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.3 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.0 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.0 | 0.3 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.0 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) |
| 0.0 | 0.0 | GO:0071879 | UDP-glucose catabolic process(GO:0006258) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.0 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.0 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.0 | 0.0 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.2 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.1 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 1.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |