Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
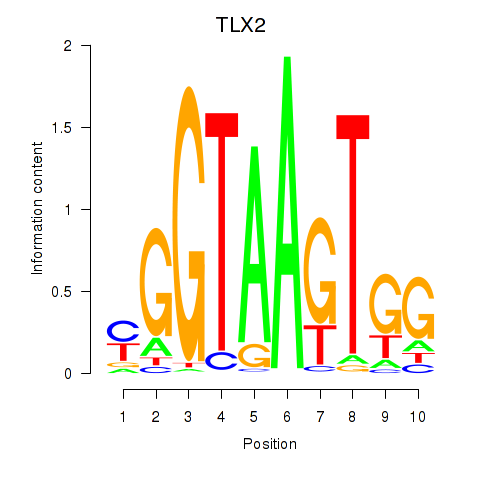
Results for TLX2
Z-value: 1.45
Transcription factors associated with TLX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TLX2
|
ENSG00000115297.9 | T cell leukemia homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TLX2 | hg19_v2_chr2_+_74741569_74741620 | -0.62 | 1.9e-01 | Click! |
Activity profile of TLX2 motif
Sorted Z-values of TLX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_948803 | 2.15 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr20_+_52824367 | 1.18 |
ENST00000371419.2
|
PFDN4
|
prefoldin subunit 4 |
| chr3_-_122283079 | 1.16 |
ENST00000471785.1
ENST00000466126.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr3_-_122283100 | 1.13 |
ENST00000492382.1
ENST00000462315.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr12_-_89918982 | 1.12 |
ENST00000549504.1
|
POC1B
|
POC1 centriolar protein B |
| chr1_-_1850697 | 1.11 |
ENST00000378598.4
ENST00000416272.1 ENST00000310991.3 |
TMEM52
|
transmembrane protein 52 |
| chr1_-_92764523 | 1.09 |
ENST00000370360.3
ENST00000534881.1 |
GLMN
|
glomulin, FKBP associated protein |
| chr7_-_120497178 | 1.00 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr9_-_80645520 | 0.92 |
ENST00000411677.1
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr7_+_130794878 | 0.91 |
ENST00000416992.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr13_+_60971427 | 0.91 |
ENST00000535286.1
ENST00000377881.2 |
TDRD3
|
tudor domain containing 3 |
| chr7_+_89783689 | 0.90 |
ENST00000297205.2
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr1_-_74663825 | 0.83 |
ENST00000370911.3
ENST00000370909.2 ENST00000354431.4 |
LRRIQ3
|
leucine-rich repeats and IQ motif containing 3 |
| chr15_+_82555169 | 0.83 |
ENST00000565432.1
ENST00000427381.2 |
FAM154B
|
family with sequence similarity 154, member B |
| chr1_+_74663994 | 0.79 |
ENST00000472069.1
|
FPGT
|
fucose-1-phosphate guanylyltransferase |
| chr14_-_54955721 | 0.78 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr15_-_34502278 | 0.77 |
ENST00000559515.1
ENST00000256544.3 ENST00000560108.1 ENST00000559462.1 |
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr7_-_33080506 | 0.76 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr15_+_82555125 | 0.74 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B
|
family with sequence similarity 154, member B |
| chrX_-_45629661 | 0.72 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr15_-_34502197 | 0.70 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr13_+_60971080 | 0.70 |
ENST00000377894.2
|
TDRD3
|
tudor domain containing 3 |
| chr3_-_149051194 | 0.67 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr3_+_122283064 | 0.65 |
ENST00000296161.4
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr3_-_182880541 | 0.62 |
ENST00000470251.1
ENST00000265598.3 |
LAMP3
|
lysosomal-associated membrane protein 3 |
| chr12_+_41221975 | 0.62 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr2_+_187371440 | 0.61 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr6_+_24775641 | 0.59 |
ENST00000378054.2
ENST00000476555.1 |
GMNN
|
geminin, DNA replication inhibitor |
| chr5_+_40841410 | 0.59 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr6_+_111303218 | 0.57 |
ENST00000441448.2
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae) |
| chr5_-_102455801 | 0.57 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr4_-_105416039 | 0.57 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr3_-_107809816 | 0.57 |
ENST00000361309.5
ENST00000355354.7 |
CD47
|
CD47 molecule |
| chr3_-_149051444 | 0.56 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr14_-_54955376 | 0.56 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr8_-_55014336 | 0.54 |
ENST00000343231.6
|
LYPLA1
|
lysophospholipase I |
| chr4_-_147443043 | 0.54 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr5_+_102455968 | 0.53 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr21_+_30671189 | 0.53 |
ENST00000286800.3
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr8_-_91657740 | 0.52 |
ENST00000422900.1
|
TMEM64
|
transmembrane protein 64 |
| chrX_-_73511908 | 0.52 |
ENST00000455395.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr10_+_35416223 | 0.52 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr12_-_27091183 | 0.51 |
ENST00000544548.1
ENST00000261191.7 ENST00000537336.1 |
ASUN
|
asunder spermatogenesis regulator |
| chr7_+_130794846 | 0.51 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr11_-_102323740 | 0.51 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr13_-_88323514 | 0.51 |
ENST00000441617.1
|
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr20_+_2795626 | 0.51 |
ENST00000603872.1
ENST00000380589.4 |
C20orf141
|
chromosome 20 open reading frame 141 |
| chr5_+_131409476 | 0.50 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr21_+_30671690 | 0.50 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr8_+_86376081 | 0.50 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr2_+_169659121 | 0.50 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr1_+_153330322 | 0.50 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr20_-_13765526 | 0.50 |
ENST00000202816.1
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr1_-_222885770 | 0.50 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr4_+_140222609 | 0.49 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr12_-_90024360 | 0.49 |
ENST00000393164.2
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr21_-_16437126 | 0.49 |
ENST00000318948.4
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr6_-_46620522 | 0.49 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr10_+_35416090 | 0.49 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr6_-_6007200 | 0.49 |
ENST00000244766.2
|
NRN1
|
neuritin 1 |
| chr4_-_147442817 | 0.48 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr11_-_22851367 | 0.48 |
ENST00000354193.4
|
SVIP
|
small VCP/p97-interacting protein |
| chr8_+_38758737 | 0.48 |
ENST00000521746.1
ENST00000420274.1 |
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr4_-_68411275 | 0.48 |
ENST00000273853.6
|
CENPC
|
centromere protein C |
| chr9_+_139847347 | 0.47 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr2_+_160568978 | 0.47 |
ENST00000409175.1
ENST00000539065.1 ENST00000259050.4 ENST00000421037.1 |
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chr12_-_99038732 | 0.47 |
ENST00000393042.3
ENST00000420861.1 ENST00000299157.4 ENST00000342502.2 |
IKBIP
|
IKBKB interacting protein |
| chr10_-_27529779 | 0.47 |
ENST00000426079.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr12_+_9144626 | 0.46 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr1_+_222886694 | 0.46 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chrX_-_77395186 | 0.46 |
ENST00000341864.5
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr7_-_22539771 | 0.45 |
ENST00000406890.2
ENST00000424363.1 |
STEAP1B
|
STEAP family member 1B |
| chr11_-_102323489 | 0.45 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr6_-_138428613 | 0.45 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector |
| chr9_-_125675576 | 0.45 |
ENST00000373659.3
|
ZBTB6
|
zinc finger and BTB domain containing 6 |
| chr11_-_93474645 | 0.44 |
ENST00000532455.1
|
TAF1D
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa |
| chr8_+_74903580 | 0.44 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr1_+_231114795 | 0.44 |
ENST00000310256.2
ENST00000366658.2 ENST00000450711.1 ENST00000435927.1 |
ARV1
|
ARV1 homolog (S. cerevisiae) |
| chr11_-_34937858 | 0.44 |
ENST00000278359.5
|
APIP
|
APAF1 interacting protein |
| chr12_-_89919220 | 0.44 |
ENST00000549035.1
ENST00000393179.4 |
POC1B
|
POC1 centriolar protein B |
| chr4_+_17812525 | 0.44 |
ENST00000251496.2
|
NCAPG
|
non-SMC condensin I complex, subunit G |
| chr4_+_39046615 | 0.43 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr9_-_80646374 | 0.43 |
ENST00000286548.4
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr4_-_165305086 | 0.43 |
ENST00000507270.1
ENST00000514618.1 ENST00000503008.1 |
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr2_-_224702201 | 0.42 |
ENST00000446015.2
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr8_-_27630102 | 0.42 |
ENST00000356537.4
ENST00000522915.1 ENST00000539095.1 |
CCDC25
|
coiled-coil domain containing 25 |
| chr1_-_222886526 | 0.42 |
ENST00000541237.1
|
AIDA
|
axin interactor, dorsalization associated |
| chr5_+_93954358 | 0.42 |
ENST00000504099.1
|
ANKRD32
|
ankyrin repeat domain 32 |
| chrX_+_49644470 | 0.41 |
ENST00000508866.2
|
USP27X
|
ubiquitin specific peptidase 27, X-linked |
| chr20_+_2795609 | 0.41 |
ENST00000554164.1
ENST00000380593.4 |
TMEM239
TMEM239
|
transmembrane protein 239 CDNA FLJ26142 fis, clone TST04526; Transmembrane protein 239; Uncharacterized protein |
| chr9_+_86595626 | 0.41 |
ENST00000445877.1
ENST00000325875.3 |
RMI1
|
RecQ mediated genome instability 1 |
| chr1_+_222885884 | 0.41 |
ENST00000340934.5
|
BROX
|
BRO1 domain and CAAX motif containing |
| chrX_-_151999269 | 0.40 |
ENST00000370277.3
|
CETN2
|
centrin, EF-hand protein, 2 |
| chr20_+_5731027 | 0.40 |
ENST00000378979.4
ENST00000303142.6 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr1_+_31769836 | 0.40 |
ENST00000344147.5
ENST00000373714.1 ENST00000546109.1 ENST00000422613.2 |
ZCCHC17
|
zinc finger, CCHC domain containing 17 |
| chr8_-_128231299 | 0.40 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr12_-_50677255 | 0.39 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr1_-_108231101 | 0.39 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr7_+_106809406 | 0.39 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr1_+_16083098 | 0.38 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr2_-_224702270 | 0.38 |
ENST00000396654.2
ENST00000396653.2 ENST00000423110.1 ENST00000443700.1 |
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr17_-_4889508 | 0.38 |
ENST00000574606.2
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr11_-_34938039 | 0.38 |
ENST00000395787.3
|
APIP
|
APAF1 interacting protein |
| chr1_+_199996733 | 0.38 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr13_-_103426081 | 0.38 |
ENST00000376022.1
ENST00000376021.4 |
TEX30
|
testis expressed 30 |
| chr3_-_179322436 | 0.38 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr7_+_77469439 | 0.37 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr5_+_94982558 | 0.37 |
ENST00000311364.4
ENST00000458310.1 |
RFESD
|
Rieske (Fe-S) domain containing |
| chr1_-_235324772 | 0.37 |
ENST00000408888.3
|
RBM34
|
RNA binding motif protein 34 |
| chr5_-_135290651 | 0.36 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr2_+_172544294 | 0.36 |
ENST00000358002.6
ENST00000435234.1 ENST00000443458.1 ENST00000412370.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr8_-_55014415 | 0.36 |
ENST00000522007.1
ENST00000521898.1 ENST00000518546.1 ENST00000316963.3 |
LYPLA1
|
lysophospholipase I |
| chr20_-_1373682 | 0.36 |
ENST00000381724.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr11_+_124543694 | 0.36 |
ENST00000227135.2
ENST00000532692.1 |
SPA17
|
sperm autoantigenic protein 17 |
| chr20_+_56884752 | 0.36 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr3_-_64253655 | 0.36 |
ENST00000498162.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr13_-_88323218 | 0.35 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr3_-_113160334 | 0.35 |
ENST00000393845.2
ENST00000295868.2 |
WDR52
|
WD repeat domain 52 |
| chr3_-_179322416 | 0.35 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr12_-_89918522 | 0.35 |
ENST00000529983.2
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chr13_-_103426112 | 0.35 |
ENST00000376032.4
ENST00000376029.3 |
TEX30
|
testis expressed 30 |
| chr3_+_148847371 | 0.34 |
ENST00000296051.2
ENST00000460120.1 |
HPS3
|
Hermansky-Pudlak syndrome 3 |
| chr11_-_85780853 | 0.34 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr15_-_71055769 | 0.34 |
ENST00000539319.1
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr1_+_24286287 | 0.34 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr9_+_72873837 | 0.34 |
ENST00000361138.5
|
SMC5
|
structural maintenance of chromosomes 5 |
| chr7_+_99971129 | 0.34 |
ENST00000394000.2
ENST00000350573.2 |
PILRA
|
paired immunoglobin-like type 2 receptor alpha |
| chr9_+_35673853 | 0.34 |
ENST00000378357.4
|
CA9
|
carbonic anhydrase IX |
| chr3_+_108308559 | 0.34 |
ENST00000486815.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr10_+_76871229 | 0.34 |
ENST00000372690.3
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr10_+_111765562 | 0.34 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr18_+_29078131 | 0.33 |
ENST00000585206.1
|
DSG2
|
desmoglein 2 |
| chrX_-_38186811 | 0.33 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr7_+_73275483 | 0.33 |
ENST00000320531.2
|
WBSCR28
|
Williams-Beuren syndrome chromosome region 28 |
| chr2_+_231921574 | 0.33 |
ENST00000308696.6
ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr14_+_54863739 | 0.32 |
ENST00000541304.1
|
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr19_-_44388116 | 0.32 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr19_+_840963 | 0.32 |
ENST00000234347.5
|
PRTN3
|
proteinase 3 |
| chr14_-_36789783 | 0.32 |
ENST00000605579.1
ENST00000604336.1 ENST00000359527.7 ENST00000603139.1 ENST00000318473.7 |
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr5_+_93954039 | 0.32 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr18_+_29077990 | 0.32 |
ENST00000261590.8
|
DSG2
|
desmoglein 2 |
| chr1_+_199996702 | 0.32 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr21_-_16437255 | 0.32 |
ENST00000400199.1
ENST00000400202.1 |
NRIP1
|
nuclear receptor interacting protein 1 |
| chr12_+_32687221 | 0.32 |
ENST00000525053.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_-_190445499 | 0.32 |
ENST00000261024.2
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr9_-_88896977 | 0.31 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr10_-_115933942 | 0.31 |
ENST00000369285.3
ENST00000369287.3 ENST00000369286.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr6_-_127664683 | 0.31 |
ENST00000528402.1
ENST00000454591.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr1_+_74663896 | 0.31 |
ENST00000370898.3
ENST00000467578.2 ENST00000370894.5 ENST00000482102.2 ENST00000609362.1 ENST00000534056.1 ENST00000557284.2 ENST00000370899.3 ENST00000370895.1 ENST00000534632.1 ENST00000370893.1 ENST00000370891.2 |
FPGT
FPGT-TNNI3K
TNNI3K
|
fucose-1-phosphate guanylyltransferase FPGT-TNNI3K readthrough TNNI3 interacting kinase |
| chr2_+_192543694 | 0.31 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr12_+_104324112 | 0.31 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr1_-_115124257 | 0.31 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr10_-_52008313 | 0.30 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr8_-_55014018 | 0.30 |
ENST00000521352.1
|
LYPLA1
|
lysophospholipase I |
| chr12_+_50794891 | 0.30 |
ENST00000517559.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr1_+_92764522 | 0.30 |
ENST00000610020.1
|
RPAP2
|
RNA polymerase II associated protein 2 |
| chr4_+_41937131 | 0.30 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr6_+_21666633 | 0.30 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr15_-_27018175 | 0.30 |
ENST00000311550.5
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr20_-_14318248 | 0.30 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr10_+_76871353 | 0.29 |
ENST00000542569.1
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr2_+_102927962 | 0.29 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr5_-_180229791 | 0.29 |
ENST00000504671.1
ENST00000507384.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr22_+_50925213 | 0.29 |
ENST00000395733.3
ENST00000216075.6 ENST00000395732.3 |
MIOX
|
myo-inositol oxygenase |
| chr16_+_66400533 | 0.29 |
ENST00000341529.3
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr3_+_48507621 | 0.29 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr3_-_143567262 | 0.29 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr3_+_108308845 | 0.29 |
ENST00000479138.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr19_-_22034809 | 0.29 |
ENST00000594012.1
ENST00000595461.1 ENST00000596899.1 |
ZNF43
|
zinc finger protein 43 |
| chr1_+_53480598 | 0.29 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr2_+_172544182 | 0.29 |
ENST00000409197.1
ENST00000456808.1 ENST00000409317.1 ENST00000409773.1 ENST00000411953.1 ENST00000409453.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr15_+_42694573 | 0.29 |
ENST00000397200.4
ENST00000569827.1 |
CAPN3
|
calpain 3, (p94) |
| chr9_+_100395891 | 0.29 |
ENST00000375147.3
|
NCBP1
|
nuclear cap binding protein subunit 1, 80kDa |
| chr6_-_127664736 | 0.29 |
ENST00000368291.2
ENST00000309620.9 ENST00000454859.3 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr8_-_105601134 | 0.28 |
ENST00000276654.5
ENST00000424843.2 |
LRP12
|
low density lipoprotein receptor-related protein 12 |
| chr9_-_125693757 | 0.28 |
ENST00000373656.3
|
ZBTB26
|
zinc finger and BTB domain containing 26 |
| chr18_+_56531584 | 0.28 |
ENST00000590287.1
|
ZNF532
|
zinc finger protein 532 |
| chr3_-_43663519 | 0.28 |
ENST00000427171.1
ENST00000292246.3 |
ANO10
|
anoctamin 10 |
| chr3_+_142315294 | 0.28 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr1_+_35734562 | 0.28 |
ENST00000314607.6
ENST00000373297.2 |
ZMYM4
|
zinc finger, MYM-type 4 |
| chr1_+_207494853 | 0.28 |
ENST00000367064.3
ENST00000367063.2 ENST00000391921.4 ENST00000367067.4 ENST00000314754.8 ENST00000367065.5 ENST00000391920.4 ENST00000367062.4 ENST00000343420.6 |
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
| chr3_-_128212016 | 0.28 |
ENST00000498200.1
ENST00000341105.2 |
GATA2
|
GATA binding protein 2 |
| chrX_+_152783131 | 0.28 |
ENST00000349466.2
ENST00000370186.1 |
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr6_-_127664475 | 0.28 |
ENST00000474289.2
ENST00000534442.1 ENST00000368289.2 ENST00000525745.1 ENST00000430841.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr12_-_95611149 | 0.28 |
ENST00000549499.1
ENST00000343958.4 ENST00000546711.1 |
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr12_+_31079652 | 0.27 |
ENST00000546076.1
ENST00000535215.1 ENST00000544427.1 ENST00000261177.9 |
TSPAN11
|
tetraspanin 11 |
| chr19_-_37019136 | 0.27 |
ENST00000592282.1
|
ZNF260
|
zinc finger protein 260 |
| chr5_-_133968459 | 0.27 |
ENST00000505758.1
ENST00000439578.1 ENST00000502286.1 |
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr5_+_102455853 | 0.27 |
ENST00000515845.1
ENST00000321521.9 ENST00000507921.1 |
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr10_-_60027642 | 0.26 |
ENST00000373935.3
|
IPMK
|
inositol polyphosphate multikinase |
| chr16_-_71598823 | 0.26 |
ENST00000566202.1
|
ZNF19
|
zinc finger protein 19 |
| chr15_+_93426514 | 0.26 |
ENST00000556722.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr10_-_73975657 | 0.26 |
ENST00000394919.1
ENST00000526751.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr17_+_42925270 | 0.26 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr16_-_15982440 | 0.26 |
ENST00000575938.1
ENST00000573396.1 ENST00000573968.1 ENST00000575744.1 ENST00000573429.1 ENST00000255759.6 ENST00000575073.1 |
FOPNL
|
FGFR1OP N-terminal like |
| chr17_-_40134339 | 0.26 |
ENST00000587727.1
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr14_-_23762777 | 0.26 |
ENST00000431326.2
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr12_+_50794730 | 0.26 |
ENST00000523389.1
ENST00000518561.1 ENST00000347328.5 ENST00000550260.1 |
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr1_+_33005020 | 0.25 |
ENST00000373510.4
ENST00000316459.4 |
ZBTB8A
|
zinc finger and BTB domain containing 8A |
| chr17_+_15604513 | 0.25 |
ENST00000481540.1
|
ZNF286A
|
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
Network of associatons between targets according to the STRING database.
First level regulatory network of TLX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.2 | 2.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 1.0 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 2.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.4 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 0.5 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.2 | 0.5 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 1.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 0.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 | 0.5 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.6 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.1 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.7 | GO:0003165 | Purkinje myocyte development(GO:0003165) |
| 0.1 | 0.5 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.4 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 1.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.3 | GO:1903414 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.1 | 1.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.5 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.7 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.3 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 1.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.5 | GO:1903061 | regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.1 | 0.4 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.3 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 0.8 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.4 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.8 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.6 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 1.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.1 | 0.6 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.3 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 0.4 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.3 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.1 | 0.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.2 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 0.1 | 0.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.1 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.1 | 0.3 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.2 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) |
| 0.1 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 1.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.9 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.1 | 0.2 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.2 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.1 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.6 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.9 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.2 | GO:0031081 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.0 | 0.2 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.3 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.3 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.6 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.3 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 1.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.0 | 0.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.5 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.3 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.2 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.5 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.6 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 1.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.3 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.5 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.5 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.3 | GO:1905216 | positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.0 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.1 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.3 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.2 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.6 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 1.6 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.0 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 1.2 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 1.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.7 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.2 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.0 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0016048 | detection of temperature stimulus(GO:0016048) sensory perception of temperature stimulus(GO:0050951) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.4 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.4 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.5 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.4 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.6 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.3 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.1 | GO:0045714 | regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.4 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.8 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.4 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.1 | 1.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.3 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.4 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.3 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.1 | 1.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 1.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 1.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.2 | 1.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 1.4 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 0.6 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.2 | 2.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 1.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.2 | 0.8 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.6 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.1 | 0.4 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.1 | 0.3 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.1 | 0.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.3 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 1.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.6 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 2.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.6 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.3 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.1 | 1.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.2 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.3 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.2 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 1.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.2 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 1.1 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0052723 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) inositol hexakisphosphate kinase activity(GO:0000828) inositol heptakisphosphate kinase activity(GO:0000829) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.3 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.4 | GO:0032407 | MutLalpha complex binding(GO:0032405) MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.3 | GO:0022821 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 1.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 1.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 2.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 1.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 2.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.2 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |