Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
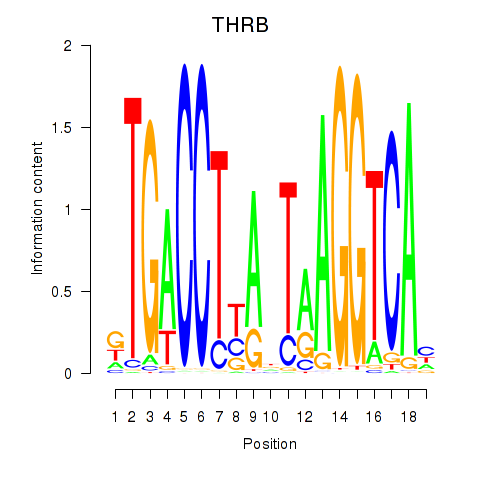
Results for THRB
Z-value: 0.43
Transcription factors associated with THRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRB
|
ENSG00000151090.13 | thyroid hormone receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| THRB | hg19_v2_chr3_-_24536453_24536500 | 0.67 | 1.5e-01 | Click! |
Activity profile of THRB motif
Sorted Z-values of THRB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_131746575 | 0.29 |
ENST00000337752.2
ENST00000378947.3 ENST00000407797.1 |
C5orf56
|
chromosome 5 open reading frame 56 |
| chr19_-_52643157 | 0.21 |
ENST00000597013.1
ENST00000600228.1 ENST00000596290.1 |
ZNF616
|
zinc finger protein 616 |
| chr17_+_7533439 | 0.18 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr17_+_1646130 | 0.17 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr20_+_3024266 | 0.16 |
ENST00000245983.2
ENST00000359100.2 ENST00000359987.1 |
GNRH2
|
gonadotropin-releasing hormone 2 |
| chr13_-_24007815 | 0.16 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr14_+_32414059 | 0.15 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr17_-_73840415 | 0.14 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr16_+_8736232 | 0.14 |
ENST00000562973.1
|
METTL22
|
methyltransferase like 22 |
| chr8_-_100905363 | 0.13 |
ENST00000524245.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr11_-_47400032 | 0.11 |
ENST00000533968.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr8_-_100905850 | 0.10 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
| chr3_+_50606901 | 0.09 |
ENST00000455834.1
|
HEMK1
|
HemK methyltransferase family member 1 |
| chr9_-_139268068 | 0.09 |
ENST00000371734.3
ENST00000371732.5 ENST00000315908.7 |
CARD9
|
caspase recruitment domain family, member 9 |
| chr16_+_72088376 | 0.09 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr11_-_64014379 | 0.07 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr4_+_189321881 | 0.07 |
ENST00000512839.1
ENST00000513313.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr8_-_100905925 | 0.06 |
ENST00000518171.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr2_-_9563319 | 0.06 |
ENST00000497105.1
ENST00000360635.3 ENST00000359712.3 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr6_-_99842041 | 0.06 |
ENST00000254759.3
ENST00000369242.1 |
COQ3
|
coenzyme Q3 methyltransferase |
| chr2_-_73869508 | 0.06 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr17_-_73839792 | 0.05 |
ENST00000590762.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr10_+_62538248 | 0.05 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1 |
| chr13_+_27844464 | 0.05 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
| chr11_-_47400062 | 0.05 |
ENST00000533030.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr12_+_54378923 | 0.05 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr11_+_73498898 | 0.05 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr21_-_37914898 | 0.04 |
ENST00000399136.1
|
CLDN14
|
claudin 14 |
| chr10_+_123970670 | 0.04 |
ENST00000496913.2
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr12_+_54378849 | 0.04 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr11_-_47399942 | 0.04 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chrX_+_101854096 | 0.04 |
ENST00000246174.2
ENST00000537008.1 ENST00000541409.1 |
ARMCX5
|
armadillo repeat containing, X-linked 5 |
| chr4_+_114214125 | 0.04 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr20_+_388679 | 0.04 |
ENST00000356286.5
ENST00000475269.1 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr12_-_21810726 | 0.03 |
ENST00000396076.1
|
LDHB
|
lactate dehydrogenase B |
| chr11_-_47400078 | 0.03 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr3_+_50606577 | 0.03 |
ENST00000434410.1
ENST00000232854.4 |
HEMK1
|
HemK methyltransferase family member 1 |
| chr12_-_21810765 | 0.03 |
ENST00000450584.1
ENST00000350669.1 |
LDHB
|
lactate dehydrogenase B |
| chr7_-_100253993 | 0.03 |
ENST00000461605.1
ENST00000160382.5 |
ACTL6B
|
actin-like 6B |
| chr7_-_38394118 | 0.02 |
ENST00000390345.2
|
TRGV4
|
T cell receptor gamma variable 4 |
| chr20_+_388791 | 0.02 |
ENST00000441733.1
ENST00000353660.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr17_-_73840774 | 0.02 |
ENST00000207549.4
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr10_+_35456444 | 0.02 |
ENST00000361599.4
|
CREM
|
cAMP responsive element modulator |
| chr18_-_47808050 | 0.02 |
ENST00000590208.1
|
MBD1
|
methyl-CpG binding domain protein 1 |
| chr8_-_124037890 | 0.02 |
ENST00000519018.1
ENST00000523036.1 |
DERL1
|
derlin 1 |
| chr10_+_62538089 | 0.02 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr1_-_113498943 | 0.02 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr11_+_46366918 | 0.01 |
ENST00000528615.1
ENST00000395574.3 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr2_-_9563216 | 0.01 |
ENST00000467606.1
ENST00000494563.1 ENST00000460001.1 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr11_+_46366799 | 0.01 |
ENST00000532868.2
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr1_-_154164534 | 0.01 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr18_-_47807829 | 0.01 |
ENST00000585672.1
ENST00000457839.2 ENST00000353909.3 ENST00000339998.6 ENST00000398493.1 |
MBD1
|
methyl-CpG binding domain protein 1 |
| chr8_+_22102626 | 0.01 |
ENST00000519237.1
ENST00000397802.4 |
POLR3D
|
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa |
| chr6_+_151042224 | 0.00 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of THRB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0044026 | DNA hypermethylation(GO:0044026) hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |