Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
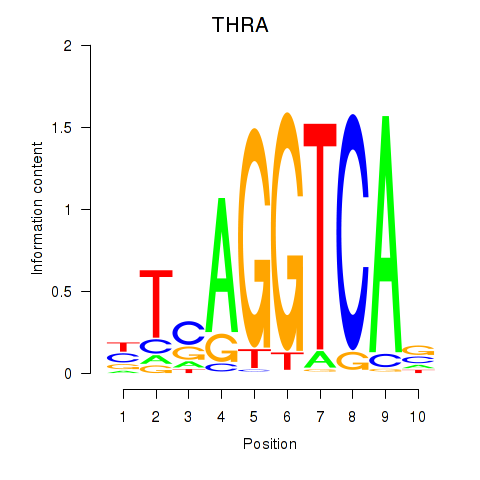
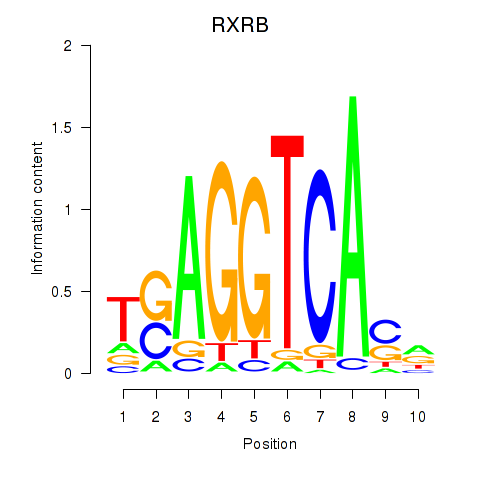
Results for THRA_RXRB
Z-value: 0.84
Transcription factors associated with THRA_RXRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRA
|
ENSG00000126351.8 | thyroid hormone receptor alpha |
|
RXRB
|
ENSG00000204231.6 | retinoid X receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| THRA | hg19_v2_chr17_+_38219063_38219154 | -0.86 | 2.9e-02 | Click! |
| RXRB | hg19_v2_chr6_-_33168391_33168465 | -0.61 | 1.9e-01 | Click! |
Activity profile of THRA_RXRB motif
Sorted Z-values of THRA_RXRB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_150693318 | 1.26 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr1_-_150693305 | 0.72 |
ENST00000368987.1
|
HORMAD1
|
HORMA domain containing 1 |
| chr5_+_162887556 | 0.63 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr12_+_102514019 | 0.60 |
ENST00000537257.1
ENST00000358383.5 ENST00000392911.2 |
PARPBP
|
PARP1 binding protein |
| chr12_+_102513950 | 0.56 |
ENST00000378128.3
ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP
|
PARP1 binding protein |
| chr6_+_151358048 | 0.52 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr9_-_33402506 | 0.48 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr8_-_99954788 | 0.48 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr12_+_41221975 | 0.47 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr2_-_55459437 | 0.47 |
ENST00000401408.1
|
CLHC1
|
clathrin heavy chain linker domain containing 1 |
| chr14_-_58893832 | 0.46 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr14_+_21492331 | 0.45 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr15_-_45422056 | 0.44 |
ENST00000267803.4
ENST00000559014.1 ENST00000558851.1 ENST00000559988.1 ENST00000558996.1 ENST00000558422.1 ENST00000559226.1 ENST00000558326.1 ENST00000558377.1 ENST00000559644.1 |
DUOXA1
|
dual oxidase maturation factor 1 |
| chr2_-_55459294 | 0.44 |
ENST00000407122.1
ENST00000406437.2 |
CLHC1
|
clathrin heavy chain linker domain containing 1 |
| chr3_+_164924716 | 0.39 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr1_+_25071848 | 0.37 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr6_-_89673280 | 0.37 |
ENST00000369475.3
ENST00000369485.4 ENST00000538899.1 ENST00000265607.6 |
RNGTT
|
RNA guanylyltransferase and 5'-phosphatase |
| chr6_+_35773070 | 0.37 |
ENST00000373853.1
ENST00000360215.1 |
LHFPL5
|
lipoma HMGIC fusion partner-like 5 |
| chr5_-_145483932 | 0.36 |
ENST00000311450.4
|
PLAC8L1
|
PLAC8-like 1 |
| chr12_+_41221794 | 0.33 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr2_+_230787201 | 0.33 |
ENST00000283946.3
|
FBXO36
|
F-box protein 36 |
| chr1_-_242612726 | 0.33 |
ENST00000459864.1
|
PLD5
|
phospholipase D family, member 5 |
| chr1_+_196621002 | 0.32 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr1_-_205719295 | 0.32 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr22_-_29137771 | 0.31 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr14_-_58893876 | 0.30 |
ENST00000555097.1
ENST00000555404.1 |
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr19_+_49109990 | 0.29 |
ENST00000321762.1
|
SPACA4
|
sperm acrosome associated 4 |
| chr1_+_196621156 | 0.29 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr12_-_88535747 | 0.29 |
ENST00000309041.7
|
CEP290
|
centrosomal protein 290kDa |
| chr6_-_150039249 | 0.27 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr16_+_85942594 | 0.27 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr13_+_76123883 | 0.26 |
ENST00000377595.3
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr19_-_56047661 | 0.26 |
ENST00000344158.3
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr22_+_17082732 | 0.25 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr6_+_64346386 | 0.25 |
ENST00000509330.1
|
PHF3
|
PHD finger protein 3 |
| chr2_-_179315786 | 0.25 |
ENST00000457633.1
ENST00000438687.3 ENST00000325748.4 |
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr1_+_178482262 | 0.24 |
ENST00000367641.3
ENST00000367639.1 |
TEX35
|
testis expressed 35 |
| chr11_-_73720122 | 0.24 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr15_+_48413211 | 0.24 |
ENST00000449382.2
|
SLC24A5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chr9_-_114246332 | 0.24 |
ENST00000602978.1
|
KIAA0368
|
KIAA0368 |
| chr2_-_179315490 | 0.24 |
ENST00000487082.1
|
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr2_-_179315453 | 0.22 |
ENST00000432031.2
|
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr12_+_56477093 | 0.22 |
ENST00000549672.1
ENST00000415288.2 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr6_-_150039170 | 0.22 |
ENST00000458696.2
ENST00000392273.3 |
LATS1
|
large tumor suppressor kinase 1 |
| chr15_+_48413169 | 0.22 |
ENST00000341459.3
ENST00000482911.2 |
SLC24A5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chr14_+_88852059 | 0.22 |
ENST00000045347.7
|
SPATA7
|
spermatogenesis associated 7 |
| chr14_-_102552659 | 0.22 |
ENST00000441629.2
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr14_-_55369525 | 0.21 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
| chr7_-_95064264 | 0.20 |
ENST00000536183.1
ENST00000433091.2 ENST00000222572.3 |
PON2
|
paraoxonase 2 |
| chr16_-_58328923 | 0.20 |
ENST00000567164.1
ENST00000219301.4 ENST00000569727.1 |
PRSS54
|
protease, serine, 54 |
| chr3_+_179322481 | 0.20 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr22_+_23243156 | 0.20 |
ENST00000390323.2
|
IGLC2
|
immunoglobulin lambda constant 2 (Kern-Oz- marker) |
| chr8_+_27632083 | 0.20 |
ENST00000519637.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr4_+_177241094 | 0.20 |
ENST00000503362.1
|
SPCS3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr16_-_58328870 | 0.19 |
ENST00000543437.1
|
PRSS54
|
protease, serine, 54 |
| chr14_+_58765103 | 0.19 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr17_-_7082668 | 0.19 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr4_+_26321284 | 0.19 |
ENST00000506956.1
ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr6_+_41196052 | 0.19 |
ENST00000341495.2
|
TREML4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr17_+_58499844 | 0.19 |
ENST00000269127.4
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chr3_+_14989186 | 0.19 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr14_+_71788096 | 0.19 |
ENST00000557151.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr11_-_17565854 | 0.18 |
ENST00000005226.7
|
USH1C
|
Usher syndrome 1C (autosomal recessive, severe) |
| chr6_+_30130969 | 0.18 |
ENST00000376694.4
|
TRIM15
|
tripartite motif containing 15 |
| chr16_-_29757272 | 0.18 |
ENST00000329410.3
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chr11_-_65837090 | 0.18 |
ENST00000529036.1
|
RP11-1167A19.2
|
Uncharacterized protein |
| chr13_+_41635617 | 0.18 |
ENST00000542082.1
|
WBP4
|
WW domain binding protein 4 |
| chr2_+_230787213 | 0.18 |
ENST00000409992.1
|
FBXO36
|
F-box protein 36 |
| chr16_-_58328884 | 0.18 |
ENST00000569079.1
|
PRSS54
|
protease, serine, 54 |
| chr20_+_56884752 | 0.18 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr3_-_98241760 | 0.17 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr12_-_102513843 | 0.17 |
ENST00000551744.2
ENST00000552283.1 |
NUP37
|
nucleoporin 37kDa |
| chr18_-_19748379 | 0.17 |
ENST00000579431.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr12_+_20963647 | 0.17 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr3_-_45957088 | 0.17 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr2_-_192016276 | 0.17 |
ENST00000413064.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr1_-_27240455 | 0.17 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr15_-_51914996 | 0.17 |
ENST00000251076.5
|
DMXL2
|
Dmx-like 2 |
| chr2_-_61389240 | 0.16 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr2_+_55459808 | 0.16 |
ENST00000404735.1
|
RPS27A
|
ribosomal protein S27a |
| chr2_-_230786679 | 0.16 |
ENST00000543084.1
ENST00000343290.5 ENST00000389044.4 ENST00000283943.5 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr4_+_26322409 | 0.16 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr1_-_91487770 | 0.16 |
ENST00000337393.5
|
ZNF644
|
zinc finger protein 644 |
| chr1_-_85156216 | 0.16 |
ENST00000342203.3
ENST00000370612.4 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr21_+_34697258 | 0.16 |
ENST00000442071.1
ENST00000442357.2 |
IFNAR1
|
interferon (alpha, beta and omega) receptor 1 |
| chr19_-_55874611 | 0.16 |
ENST00000424985.3
|
FAM71E2
|
family with sequence similarity 71, member E2 |
| chr12_-_80328949 | 0.16 |
ENST00000450142.2
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr7_+_5085452 | 0.15 |
ENST00000353796.3
ENST00000396912.1 ENST00000396904.2 |
RBAK
RBAK-RBAKDN
|
RB-associated KRAB zinc finger RBAK-RBAKDN readthrough |
| chr2_+_85661918 | 0.15 |
ENST00000340326.2
|
SH2D6
|
SH2 domain containing 6 |
| chr14_-_82000140 | 0.15 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr7_-_108097144 | 0.15 |
ENST00000418239.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr3_-_179322416 | 0.15 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr17_-_41132088 | 0.15 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr10_+_26986582 | 0.14 |
ENST00000376215.5
ENST00000376203.5 |
PDSS1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr11_-_118661828 | 0.14 |
ENST00000264018.4
|
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr14_+_58765305 | 0.14 |
ENST00000445108.1
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr11_-_73720276 | 0.14 |
ENST00000348534.4
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr5_+_145583156 | 0.14 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr8_+_104033296 | 0.14 |
ENST00000521514.1
ENST00000518738.1 |
ATP6V1C1
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 |
| chr18_-_268019 | 0.14 |
ENST00000261600.6
|
THOC1
|
THO complex 1 |
| chr17_-_38821373 | 0.14 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr19_+_32836499 | 0.14 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr1_-_203144941 | 0.14 |
ENST00000255416.4
|
MYBPH
|
myosin binding protein H |
| chr9_-_72374848 | 0.14 |
ENST00000377200.5
ENST00000340434.4 ENST00000472967.2 |
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1 |
| chr3_+_87276407 | 0.14 |
ENST00000471660.1
ENST00000263780.4 ENST00000494980.1 |
CHMP2B
|
charged multivesicular body protein 2B |
| chr14_+_57857262 | 0.14 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr7_+_94536514 | 0.14 |
ENST00000413325.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr3_-_179322436 | 0.14 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr22_-_30661807 | 0.14 |
ENST00000403389.1
|
OSM
|
oncostatin M |
| chr2_+_55459495 | 0.14 |
ENST00000272317.6
ENST00000449323.1 |
RPS27A
|
ribosomal protein S27a |
| chr6_+_139456226 | 0.14 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr22_-_20138302 | 0.13 |
ENST00000540078.1
ENST00000439765.2 |
AC006547.14
|
uncharacterized protein LOC388849 |
| chr12_+_20963632 | 0.13 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr11_-_62437486 | 0.13 |
ENST00000528115.1
|
C11orf48
|
chromosome 11 open reading frame 48 |
| chrX_+_114827818 | 0.13 |
ENST00000420625.2
|
PLS3
|
plastin 3 |
| chr2_+_228190066 | 0.13 |
ENST00000436237.1
ENST00000443428.2 ENST00000418961.1 |
MFF
|
mitochondrial fission factor |
| chr3_-_47205066 | 0.13 |
ENST00000412450.1
|
SETD2
|
SET domain containing 2 |
| chr17_+_60457914 | 0.13 |
ENST00000305286.3
ENST00000520404.1 ENST00000518576.1 |
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr8_-_53626974 | 0.13 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr8_-_42698292 | 0.13 |
ENST00000529779.1
|
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr12_+_27849378 | 0.13 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr7_+_94536898 | 0.13 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr17_+_48046671 | 0.13 |
ENST00000505318.2
|
DLX4
|
distal-less homeobox 4 |
| chr2_+_65663812 | 0.12 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr9_+_36572851 | 0.12 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr1_-_85040090 | 0.12 |
ENST00000370630.5
|
CTBS
|
chitobiase, di-N-acetyl- |
| chr4_+_26165074 | 0.12 |
ENST00000512351.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr2_-_230786619 | 0.12 |
ENST00000389045.3
ENST00000409677.1 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr10_+_64893039 | 0.12 |
ENST00000277746.6
ENST00000435510.2 |
NRBF2
|
nuclear receptor binding factor 2 |
| chr5_+_112849373 | 0.12 |
ENST00000161863.4
ENST00000515883.1 |
YTHDC2
|
YTH domain containing 2 |
| chr2_+_219745020 | 0.12 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr12_+_95611569 | 0.12 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr17_-_58499766 | 0.12 |
ENST00000588898.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr8_+_27632047 | 0.12 |
ENST00000397418.2
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr8_+_63161550 | 0.12 |
ENST00000328472.5
|
NKAIN3
|
Na+/K+ transporting ATPase interacting 3 |
| chr9_+_131452239 | 0.12 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr2_-_165698662 | 0.12 |
ENST00000194871.6
ENST00000445474.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr3_+_179322573 | 0.11 |
ENST00000493866.1
ENST00000472629.1 ENST00000482604.1 |
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr6_+_41196077 | 0.11 |
ENST00000448827.2
|
TREML4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr14_-_31889782 | 0.11 |
ENST00000543095.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chr5_+_66124590 | 0.11 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_82165350 | 0.11 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chr1_+_70876926 | 0.11 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr14_+_88851874 | 0.11 |
ENST00000393545.4
ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7
|
spermatogenesis associated 7 |
| chr19_+_57862622 | 0.11 |
ENST00000391705.3
ENST00000443917.2 ENST00000598744.1 |
ZNF304
|
zinc finger protein 304 |
| chr1_-_173886491 | 0.11 |
ENST00000367698.3
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chrX_+_114827851 | 0.11 |
ENST00000539310.1
|
PLS3
|
plastin 3 |
| chr3_-_98241713 | 0.10 |
ENST00000502288.1
ENST00000512147.1 ENST00000510541.1 ENST00000503621.1 ENST00000511081.1 |
CLDND1
|
claudin domain containing 1 |
| chrX_+_67913471 | 0.10 |
ENST00000374597.3
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr6_-_31745037 | 0.10 |
ENST00000375688.4
|
VWA7
|
von Willebrand factor A domain containing 7 |
| chr5_+_145583107 | 0.10 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr15_-_34630234 | 0.10 |
ENST00000558667.1
ENST00000561120.1 ENST00000559236.1 ENST00000397702.2 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr8_-_18666360 | 0.10 |
ENST00000286485.8
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr14_-_104028595 | 0.10 |
ENST00000337322.4
ENST00000445922.2 |
BAG5
|
BCL2-associated athanogene 5 |
| chr3_+_141105235 | 0.10 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr8_-_42698433 | 0.10 |
ENST00000345117.2
ENST00000254250.3 |
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr9_+_84304628 | 0.10 |
ENST00000437181.1
|
RP11-154D17.1
|
RP11-154D17.1 |
| chr15_+_45422131 | 0.10 |
ENST00000321429.4
|
DUOX1
|
dual oxidase 1 |
| chr14_-_31889737 | 0.10 |
ENST00000382464.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chr3_-_50340996 | 0.10 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr19_+_38880695 | 0.10 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr18_+_32558380 | 0.09 |
ENST00000588349.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr9_-_88897426 | 0.09 |
ENST00000375991.4
ENST00000326094.4 |
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr22_-_29138386 | 0.09 |
ENST00000544772.1
|
CHEK2
|
checkpoint kinase 2 |
| chr17_+_48046538 | 0.09 |
ENST00000240306.3
|
DLX4
|
distal-less homeobox 4 |
| chr2_-_68479614 | 0.09 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr13_-_76111945 | 0.09 |
ENST00000355801.4
ENST00000406936.3 |
COMMD6
|
COMM domain containing 6 |
| chr6_-_31745085 | 0.09 |
ENST00000375686.3
ENST00000447450.1 |
VWA7
|
von Willebrand factor A domain containing 7 |
| chr3_-_98241598 | 0.09 |
ENST00000513287.1
ENST00000514537.1 ENST00000508071.1 ENST00000507944.1 |
CLDND1
|
claudin domain containing 1 |
| chr1_-_103574024 | 0.09 |
ENST00000512756.1
ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1
|
collagen, type XI, alpha 1 |
| chr12_-_25102252 | 0.09 |
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr3_-_197024965 | 0.09 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr5_-_158636512 | 0.09 |
ENST00000424310.2
|
RNF145
|
ring finger protein 145 |
| chr12_+_95612006 | 0.09 |
ENST00000551311.1
ENST00000546445.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr1_+_17634689 | 0.09 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr15_-_34629922 | 0.09 |
ENST00000559484.1
ENST00000354181.3 ENST00000558589.1 ENST00000458406.2 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr14_-_21492113 | 0.09 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr17_-_15496722 | 0.09 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr1_-_109940550 | 0.08 |
ENST00000256637.6
|
SORT1
|
sortilin 1 |
| chr5_+_170288856 | 0.08 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr3_-_126236588 | 0.08 |
ENST00000383579.3
|
UROC1
|
urocanate hydratase 1 |
| chr11_-_10920714 | 0.08 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr3_-_195270162 | 0.08 |
ENST00000438848.1
ENST00000328432.3 |
PPP1R2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr11_+_57310114 | 0.08 |
ENST00000527972.1
ENST00000399154.2 |
SMTNL1
|
smoothelin-like 1 |
| chr12_+_120031264 | 0.08 |
ENST00000426426.1
|
TMEM233
|
transmembrane protein 233 |
| chr7_-_151433393 | 0.08 |
ENST00000492843.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr10_-_33625154 | 0.08 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr15_-_83315874 | 0.08 |
ENST00000569257.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr19_-_45826125 | 0.08 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr12_-_96184533 | 0.08 |
ENST00000343702.4
ENST00000344911.4 |
NTN4
|
netrin 4 |
| chr4_+_48343339 | 0.07 |
ENST00000264313.6
|
SLAIN2
|
SLAIN motif family, member 2 |
| chr2_+_86426478 | 0.07 |
ENST00000254644.8
ENST00000605125.1 ENST00000337109.4 ENST00000409180.1 |
MRPL35
|
mitochondrial ribosomal protein L35 |
| chr1_+_70876891 | 0.07 |
ENST00000411986.2
|
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr19_+_49467232 | 0.07 |
ENST00000599784.1
ENST00000594305.1 |
CTD-2639E6.9
|
CTD-2639E6.9 |
| chr2_-_33824382 | 0.07 |
ENST00000238823.8
|
FAM98A
|
family with sequence similarity 98, member A |
| chr2_-_100106419 | 0.07 |
ENST00000393445.3
ENST00000258428.3 |
REV1
|
REV1, polymerase (DNA directed) |
| chr4_-_54457783 | 0.07 |
ENST00000263925.7
ENST00000512247.1 |
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr1_-_155880672 | 0.07 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr11_+_111896090 | 0.07 |
ENST00000393051.1
|
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr3_+_12330560 | 0.07 |
ENST00000397026.2
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
Network of associatons between targets according to the STRING database.
First level regulatory network of THRA_RXRB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 0.1 | 0.4 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.4 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 0.3 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.5 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.5 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 1.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.5 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.3 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 0.1 | GO:0038194 | thyroid-stimulating hormone signaling pathway(GO:0038194) |
| 0.1 | 0.5 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.2 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.4 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.1 | 0.2 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.0 | 0.7 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.5 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.2 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.5 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.4 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.0 | 0.3 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.3 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) kidney rudiment formation(GO:0072003) |
| 0.0 | 0.1 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.0 | 0.1 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.0 | 0.1 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.3 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.9 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.5 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.0 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.7 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 2.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) glial limiting end-foot(GO:0097451) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0035838 | growing cell tip(GO:0035838) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.1 | 0.5 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.2 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.7 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.2 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |