Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
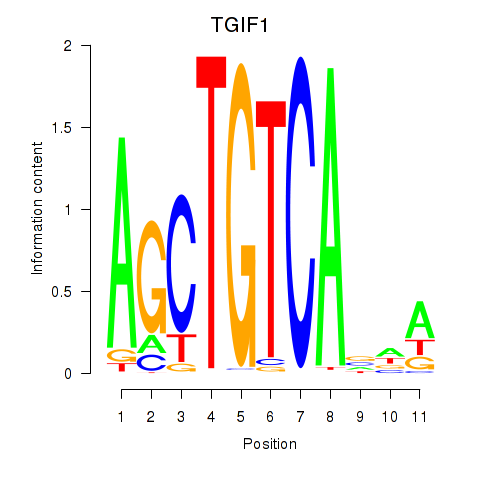
Results for TGIF1
Z-value: 0.80
Transcription factors associated with TGIF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TGIF1
|
ENSG00000177426.16 | TGFB induced factor homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF1 | hg19_v2_chr18_+_3449695_3449712 | -0.77 | 7.4e-02 | Click! |
Activity profile of TGIF1 motif
Sorted Z-values of TGIF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_25348186 | 0.57 |
ENST00000555711.1
ENST00000556885.1 ENST00000554266.1 ENST00000556351.1 ENST00000556927.1 ENST00000556402.1 ENST00000553788.1 |
LYRM5
|
LYR motif containing 5 |
| chr6_+_80714332 | 0.53 |
ENST00000502580.1
ENST00000511260.1 |
TTK
|
TTK protein kinase |
| chr8_+_71485681 | 0.48 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr8_+_72755367 | 0.48 |
ENST00000537896.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr20_-_31124186 | 0.44 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr1_-_65533390 | 0.38 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr6_+_131521120 | 0.38 |
ENST00000537868.1
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr1_+_95616933 | 0.37 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr5_-_66942617 | 0.37 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5 |
| chr6_+_80714318 | 0.37 |
ENST00000369798.2
|
TTK
|
TTK protein kinase |
| chr12_+_25348139 | 0.35 |
ENST00000557540.2
ENST00000381356.4 |
LYRM5
|
LYR motif containing 5 |
| chr2_+_37311588 | 0.34 |
ENST00000409774.1
ENST00000608836.1 |
GPATCH11
|
G patch domain containing 11 |
| chr18_-_54318353 | 0.31 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr1_-_193075180 | 0.31 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr6_-_97731019 | 0.28 |
ENST00000275053.4
|
MMS22L
|
MMS22-like, DNA repair protein |
| chr17_-_66287350 | 0.26 |
ENST00000580666.1
ENST00000583477.1 |
SLC16A6
|
solute carrier family 16, member 6 |
| chr14_+_31494672 | 0.26 |
ENST00000542754.2
ENST00000313566.6 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr10_+_94608245 | 0.25 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr2_+_37311645 | 0.25 |
ENST00000281932.5
|
GPATCH11
|
G patch domain containing 11 |
| chr7_+_95401877 | 0.25 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr10_+_98592674 | 0.24 |
ENST00000356016.3
ENST00000371097.4 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr4_-_175443943 | 0.24 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr2_+_118846008 | 0.24 |
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2 |
| chr2_-_179914760 | 0.23 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr22_+_17082732 | 0.23 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr3_+_122920847 | 0.23 |
ENST00000466519.1
ENST00000480631.1 ENST00000491366.1 ENST00000487572.1 |
SEC22A
|
SEC22 vesicle trafficking protein homolog A (S. cerevisiae) |
| chr1_-_220608023 | 0.23 |
ENST00000416839.2
|
AC096644.1
|
Uncharacterized protein |
| chr3_-_101232019 | 0.23 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr1_+_150039877 | 0.22 |
ENST00000419023.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr3_+_112280857 | 0.22 |
ENST00000492406.1
ENST00000468642.1 |
SLC35A5
|
solute carrier family 35, member A5 |
| chr12_+_62654155 | 0.22 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr7_-_97501469 | 0.22 |
ENST00000414884.1
ENST00000442734.1 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr15_-_81195510 | 0.22 |
ENST00000561295.1
|
RP11-351M8.1
|
Uncharacterized protein |
| chr10_-_75255668 | 0.22 |
ENST00000545874.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr12_+_86268065 | 0.22 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chr4_-_83483094 | 0.21 |
ENST00000508701.1
ENST00000454948.3 |
TMEM150C
|
transmembrane protein 150C |
| chr11_-_8954491 | 0.21 |
ENST00000526227.1
ENST00000525780.1 ENST00000326053.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr6_-_18249971 | 0.21 |
ENST00000507591.1
|
DEK
|
DEK oncogene |
| chr14_+_21492331 | 0.20 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr2_+_58134756 | 0.20 |
ENST00000435505.2
ENST00000417641.2 |
VRK2
|
vaccinia related kinase 2 |
| chr12_-_25348007 | 0.20 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr3_-_142166796 | 0.20 |
ENST00000392981.2
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr9_-_80645520 | 0.20 |
ENST00000411677.1
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr7_+_66800928 | 0.20 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr17_+_74734052 | 0.19 |
ENST00000590514.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr17_-_66287257 | 0.19 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr12_-_76461249 | 0.18 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr3_+_63897605 | 0.18 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr3_-_142166904 | 0.18 |
ENST00000264951.4
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr5_+_157170703 | 0.17 |
ENST00000286307.5
|
LSM11
|
LSM11, U7 small nuclear RNA associated |
| chr2_+_29353520 | 0.17 |
ENST00000438819.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr19_-_36643329 | 0.17 |
ENST00000589154.1
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr3_-_142166846 | 0.17 |
ENST00000463916.1
ENST00000544157.1 |
XRN1
|
5'-3' exoribonuclease 1 |
| chr1_-_241799232 | 0.17 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr9_-_100459639 | 0.17 |
ENST00000375128.4
|
XPA
|
xeroderma pigmentosum, complementation group A |
| chr7_-_50628745 | 0.16 |
ENST00000380984.4
ENST00000357936.5 ENST00000426377.1 |
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr5_-_94620239 | 0.16 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr8_-_128960591 | 0.16 |
ENST00000539634.1
|
TMEM75
|
transmembrane protein 75 |
| chr11_-_111781554 | 0.16 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr14_+_90864504 | 0.16 |
ENST00000544280.2
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr1_+_231114795 | 0.16 |
ENST00000310256.2
ENST00000366658.2 ENST00000450711.1 ENST00000435927.1 |
ARV1
|
ARV1 homolog (S. cerevisiae) |
| chr3_-_52719912 | 0.16 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr9_+_125273081 | 0.16 |
ENST00000335302.5
|
OR1J2
|
olfactory receptor, family 1, subfamily J, member 2 |
| chr4_-_16228120 | 0.16 |
ENST00000405303.2
|
TAPT1
|
transmembrane anterior posterior transformation 1 |
| chr6_-_79787902 | 0.16 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr1_-_150669500 | 0.16 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr3_+_122920767 | 0.16 |
ENST00000492595.1
ENST00000473494.2 ENST00000481965.2 |
SEC22A
|
SEC22 vesicle trafficking protein homolog A (S. cerevisiae) |
| chr5_+_139505520 | 0.16 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr5_+_72143988 | 0.15 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr4_+_55095428 | 0.15 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr6_+_155334780 | 0.15 |
ENST00000538270.1
ENST00000535231.1 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr6_+_119215308 | 0.15 |
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr18_-_54305658 | 0.15 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chrX_+_155110956 | 0.15 |
ENST00000286448.6
ENST00000262640.6 ENST00000460621.1 |
VAMP7
|
vesicle-associated membrane protein 7 |
| chr3_-_57530051 | 0.15 |
ENST00000311202.6
ENST00000351747.2 ENST00000495027.1 ENST00000389536.4 |
DNAH12
|
dynein, axonemal, heavy chain 12 |
| chr3_+_169491171 | 0.14 |
ENST00000356716.4
|
MYNN
|
myoneurin |
| chr8_+_99076750 | 0.14 |
ENST00000545282.1
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr3_-_52719888 | 0.14 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr19_-_38085633 | 0.14 |
ENST00000593133.1
ENST00000590751.1 ENST00000358744.3 ENST00000328550.2 ENST00000451802.2 |
ZNF571
|
zinc finger protein 571 |
| chr12_+_19593515 | 0.14 |
ENST00000360995.4
|
AEBP2
|
AE binding protein 2 |
| chr4_-_89205879 | 0.14 |
ENST00000608933.1
ENST00000315194.4 ENST00000514204.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr21_+_25801041 | 0.14 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr17_-_39341594 | 0.14 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr5_+_40841410 | 0.14 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr3_+_160473343 | 0.14 |
ENST00000497343.1
|
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr13_+_41885341 | 0.14 |
ENST00000379406.3
ENST00000379367.3 ENST00000403412.3 |
NAA16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr12_+_40618873 | 0.14 |
ENST00000298910.7
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr2_+_223916862 | 0.14 |
ENST00000604125.1
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4 |
| chr18_-_73967160 | 0.14 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr10_+_94352956 | 0.14 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr2_-_201936302 | 0.14 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr4_+_52709229 | 0.13 |
ENST00000334635.5
ENST00000381441.3 ENST00000381437.4 |
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 |
| chrX_+_133930798 | 0.13 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr7_-_156685841 | 0.13 |
ENST00000354505.4
ENST00000540390.1 |
LMBR1
|
limb development membrane protein 1 |
| chr2_-_201729284 | 0.13 |
ENST00000434813.2
|
CLK1
|
CDC-like kinase 1 |
| chr4_-_105416039 | 0.13 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr3_-_185641681 | 0.13 |
ENST00000259043.7
|
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr16_+_2880254 | 0.13 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B |
| chr13_-_96329048 | 0.13 |
ENST00000606011.1
ENST00000499499.2 |
DNAJC3-AS1
|
DNAJC3 antisense RNA 1 (head to head) |
| chr11_+_60552797 | 0.13 |
ENST00000308287.1
|
MS4A10
|
membrane-spanning 4-domains, subfamily A, member 10 |
| chr15_+_93426514 | 0.13 |
ENST00000556722.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr8_+_38244638 | 0.12 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chrX_-_134429952 | 0.12 |
ENST00000370764.1
|
ZNF75D
|
zinc finger protein 75D |
| chr1_-_204436344 | 0.12 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr1_+_150039787 | 0.12 |
ENST00000535106.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr15_-_70994612 | 0.12 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr4_+_146402843 | 0.12 |
ENST00000514831.1
|
SMAD1
|
SMAD family member 1 |
| chrX_+_67718863 | 0.12 |
ENST00000374622.2
|
YIPF6
|
Yip1 domain family, member 6 |
| chr6_-_127663543 | 0.12 |
ENST00000531582.1
|
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr1_+_6615241 | 0.12 |
ENST00000333172.6
ENST00000328191.4 ENST00000351136.3 |
TAS1R1
|
taste receptor, type 1, member 1 |
| chr11_-_111781454 | 0.12 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
| chr4_-_16228083 | 0.12 |
ENST00000399920.3
|
TAPT1
|
transmembrane anterior posterior transformation 1 |
| chr8_-_67090825 | 0.12 |
ENST00000276571.3
|
CRH
|
corticotropin releasing hormone |
| chr13_+_96329381 | 0.12 |
ENST00000602402.1
ENST00000376795.6 |
DNAJC3
|
DnaJ (Hsp40) homolog, subfamily C, member 3 |
| chr14_+_73563735 | 0.12 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr1_-_220445757 | 0.11 |
ENST00000358951.2
|
RAB3GAP2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr3_+_186692745 | 0.11 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr1_-_33168336 | 0.11 |
ENST00000373484.3
|
SYNC
|
syncoilin, intermediate filament protein |
| chr8_+_31497271 | 0.11 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr1_+_111889212 | 0.11 |
ENST00000369737.4
|
PIFO
|
primary cilia formation |
| chr5_+_141348598 | 0.11 |
ENST00000394520.2
ENST00000347642.3 |
RNF14
|
ring finger protein 14 |
| chr1_+_228395755 | 0.11 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr10_+_60028818 | 0.11 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr15_+_42841008 | 0.11 |
ENST00000260372.3
ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2
|
HAUS augmin-like complex, subunit 2 |
| chr15_-_66084621 | 0.11 |
ENST00000564674.1
|
DENND4A
|
DENN/MADD domain containing 4A |
| chr10_+_99349450 | 0.11 |
ENST00000370640.3
|
C10orf62
|
chromosome 10 open reading frame 62 |
| chr13_+_78109804 | 0.11 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr13_+_33160553 | 0.11 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr3_+_188817891 | 0.11 |
ENST00000412373.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr7_-_94285472 | 0.11 |
ENST00000437425.2
ENST00000447873.1 ENST00000415788.2 |
SGCE
|
sarcoglycan, epsilon |
| chr2_-_201729393 | 0.11 |
ENST00000321356.4
|
CLK1
|
CDC-like kinase 1 |
| chr1_+_161691353 | 0.11 |
ENST00000367948.2
|
FCRLB
|
Fc receptor-like B |
| chr15_+_75335604 | 0.10 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr9_-_35812236 | 0.10 |
ENST00000340291.2
|
SPAG8
|
sperm associated antigen 8 |
| chr3_-_3221358 | 0.10 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr17_-_57232596 | 0.10 |
ENST00000581068.1
ENST00000330137.7 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr17_-_57232525 | 0.10 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr2_-_228579305 | 0.10 |
ENST00000456524.1
|
SLC19A3
|
solute carrier family 19 (thiamine transporter), member 3 |
| chr5_+_32788945 | 0.10 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr7_+_23637118 | 0.10 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr9_+_17134980 | 0.10 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chrM_+_5824 | 0.10 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr16_+_50776021 | 0.10 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr1_-_150979333 | 0.10 |
ENST00000312210.5
|
FAM63A
|
family with sequence similarity 63, member A |
| chr10_-_124768300 | 0.10 |
ENST00000368886.5
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus) |
| chr12_+_62654119 | 0.10 |
ENST00000353364.3
ENST00000549523.1 ENST00000280377.5 |
USP15
|
ubiquitin specific peptidase 15 |
| chr12_-_4647606 | 0.09 |
ENST00000261250.3
ENST00000541014.1 ENST00000545746.1 ENST00000542080.1 |
C12orf4
|
chromosome 12 open reading frame 4 |
| chr9_+_17135016 | 0.09 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr4_+_186990298 | 0.09 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr2_+_27505260 | 0.09 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr4_-_175443788 | 0.09 |
ENST00000541923.1
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr3_-_123168551 | 0.09 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr8_+_26240414 | 0.09 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr6_+_109416684 | 0.09 |
ENST00000521522.1
ENST00000524064.1 ENST00000522608.1 ENST00000521503.1 ENST00000519407.1 ENST00000519095.1 ENST00000368968.2 ENST00000522490.1 ENST00000523209.1 ENST00000368970.2 ENST00000520883.1 ENST00000523787.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr16_+_53088885 | 0.09 |
ENST00000566029.1
ENST00000447540.1 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr17_-_34122596 | 0.09 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr20_-_50418972 | 0.09 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr14_+_31494841 | 0.09 |
ENST00000556232.1
ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr2_-_71454185 | 0.09 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr5_+_133984462 | 0.09 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr1_+_22379179 | 0.09 |
ENST00000315554.8
ENST00000421089.2 |
CDC42
|
cell division cycle 42 |
| chr1_+_240177627 | 0.09 |
ENST00000447095.1
|
FMN2
|
formin 2 |
| chr7_-_94285402 | 0.09 |
ENST00000428696.2
ENST00000445866.2 |
SGCE
|
sarcoglycan, epsilon |
| chr8_+_94929077 | 0.09 |
ENST00000297598.4
ENST00000520614.1 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr1_+_100111580 | 0.09 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr7_-_94285511 | 0.09 |
ENST00000265735.7
|
SGCE
|
sarcoglycan, epsilon |
| chr6_+_17600576 | 0.09 |
ENST00000259963.3
|
FAM8A1
|
family with sequence similarity 8, member A1 |
| chr8_+_38243721 | 0.09 |
ENST00000527334.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr6_+_83777374 | 0.08 |
ENST00000349129.2
ENST00000237163.5 ENST00000536812.1 |
DOPEY1
|
dopey family member 1 |
| chr1_-_247275719 | 0.08 |
ENST00000408893.2
|
C1orf229
|
chromosome 1 open reading frame 229 |
| chr7_-_100493482 | 0.08 |
ENST00000411582.1
ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr5_-_159797627 | 0.08 |
ENST00000393975.3
|
C1QTNF2
|
C1q and tumor necrosis factor related protein 2 |
| chr3_-_180707466 | 0.08 |
ENST00000491873.1
ENST00000486355.1 ENST00000382564.2 |
DNAJC19
|
DnaJ (Hsp40) homolog, subfamily C, member 19 |
| chr10_-_21806759 | 0.08 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr1_+_186798073 | 0.08 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr10_+_24528108 | 0.08 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr14_-_65569244 | 0.08 |
ENST00000557277.1
ENST00000556892.1 |
MAX
|
MYC associated factor X |
| chr13_-_48575376 | 0.08 |
ENST00000434484.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr3_+_156807663 | 0.08 |
ENST00000467995.1
ENST00000474477.1 ENST00000471719.1 |
LINC00881
|
long intergenic non-protein coding RNA 881 |
| chr15_-_66084428 | 0.08 |
ENST00000443035.3
ENST00000431932.2 |
DENND4A
|
DENN/MADD domain containing 4A |
| chr11_-_111781610 | 0.08 |
ENST00000525823.1
|
CRYAB
|
crystallin, alpha B |
| chr16_+_31366455 | 0.08 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr13_+_50589390 | 0.08 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr12_+_49687425 | 0.08 |
ENST00000257860.4
|
PRPH
|
peripherin |
| chr3_+_69788576 | 0.08 |
ENST00000352241.4
ENST00000448226.2 |
MITF
|
microphthalmia-associated transcription factor |
| chr14_-_24732368 | 0.08 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1 |
| chr6_+_24775153 | 0.08 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr1_-_247335269 | 0.08 |
ENST00000543802.2
ENST00000491356.1 ENST00000472531.1 ENST00000340684.6 |
ZNF124
|
zinc finger protein 124 |
| chr4_+_128886584 | 0.08 |
ENST00000513371.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr12_-_109027643 | 0.08 |
ENST00000388962.3
ENST00000550948.1 |
SELPLG
|
selectin P ligand |
| chr14_-_24732403 | 0.08 |
ENST00000206765.6
|
TGM1
|
transglutaminase 1 |
| chr2_-_73460334 | 0.08 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr22_+_41347363 | 0.08 |
ENST00000216225.8
|
RBX1
|
ring-box 1, E3 ubiquitin protein ligase |
| chr1_+_166808692 | 0.08 |
ENST00000367876.4
|
POGK
|
pogo transposable element with KRAB domain |
| chr12_+_59989918 | 0.08 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr17_+_61571746 | 0.08 |
ENST00000579409.1
|
ACE
|
angiotensin I converting enzyme |
| chr12_+_40618764 | 0.08 |
ENST00000343742.2
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr1_+_111888890 | 0.08 |
ENST00000369738.4
|
PIFO
|
primary cilia formation |
| chr12_-_39837192 | 0.07 |
ENST00000361961.3
ENST00000395670.3 |
KIF21A
|
kinesin family member 21A |
| chr5_-_130868688 | 0.07 |
ENST00000504575.1
ENST00000513227.1 |
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TGIF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.5 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.2 | GO:1902823 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 0.2 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.0 | 0.2 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.4 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:1903371 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.2 | GO:0070981 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.3 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0034343 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.3 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.0 | 0.2 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.0 | GO:0008052 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0090135 | positive regulation of synapse structural plasticity(GO:0051835) actin filament branching(GO:0090135) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0045213 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.1 | GO:0050777 | negative regulation of immune response(GO:0050777) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.3 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.2 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.2 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.3 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.1 | 0.2 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 0.5 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.3 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.0 | 0.2 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0031768 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |