Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
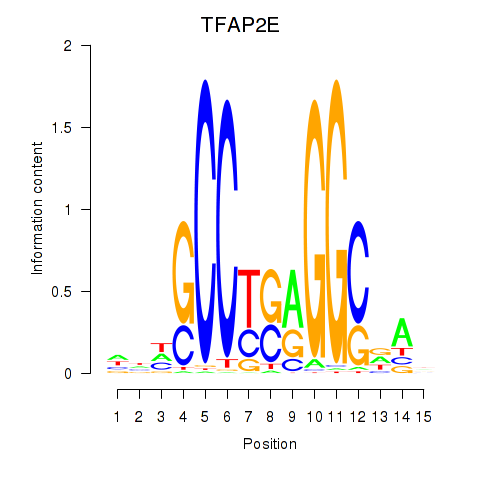
Results for TFAP2E
Z-value: 0.40
Transcription factors associated with TFAP2E
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2E
|
ENSG00000116819.6 | transcription factor AP-2 epsilon |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFAP2E | hg19_v2_chr1_+_36038971_36038971 | 0.49 | 3.2e-01 | Click! |
Activity profile of TFAP2E motif
Sorted Z-values of TFAP2E motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_170459154 | 0.38 |
ENST00000473110.1
|
RP11-373E16.4
|
RP11-373E16.4 |
| chr20_-_30539773 | 0.23 |
ENST00000202017.4
|
PDRG1
|
p53 and DNA-damage regulated 1 |
| chr6_+_13272904 | 0.22 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr22_+_22676808 | 0.21 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr19_+_54960358 | 0.21 |
ENST00000439657.1
ENST00000376514.2 ENST00000376526.4 ENST00000436479.1 |
LENG8
|
leukocyte receptor cluster (LRC) member 8 |
| chr19_-_39264072 | 0.21 |
ENST00000599035.1
ENST00000378626.4 |
LGALS7
|
lectin, galactoside-binding, soluble, 7 |
| chr19_+_54960790 | 0.21 |
ENST00000443957.1
|
LENG8
|
leukocyte receptor cluster (LRC) member 8 |
| chr19_+_3572758 | 0.19 |
ENST00000416526.1
|
HMG20B
|
high mobility group 20B |
| chr4_-_82393052 | 0.19 |
ENST00000335927.7
ENST00000504863.1 ENST00000264400.2 |
RASGEF1B
|
RasGEF domain family, member 1B |
| chr19_-_11039188 | 0.18 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr2_+_38893208 | 0.16 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr19_-_11039261 | 0.15 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr19_-_54676846 | 0.14 |
ENST00000301187.4
|
TMC4
|
transmembrane channel-like 4 |
| chr19_+_11039391 | 0.14 |
ENST00000270502.6
|
C19orf52
|
chromosome 19 open reading frame 52 |
| chr7_+_100464760 | 0.14 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr17_+_72428218 | 0.13 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_-_66056596 | 0.13 |
ENST00000471387.2
ENST00000359461.6 ENST00000376901.4 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr11_+_118478313 | 0.13 |
ENST00000356063.5
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1 |
| chr15_+_23810853 | 0.12 |
ENST00000568252.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr2_-_133429091 | 0.12 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr4_-_85419603 | 0.12 |
ENST00000295886.4
|
NKX6-1
|
NK6 homeobox 1 |
| chr2_+_239335449 | 0.12 |
ENST00000264607.4
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr17_+_36584662 | 0.12 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr16_-_28506840 | 0.11 |
ENST00000569430.1
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr11_-_65625014 | 0.11 |
ENST00000534784.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr6_+_292253 | 0.11 |
ENST00000603453.1
ENST00000605315.1 ENST00000603881.1 |
DUSP22
|
dual specificity phosphatase 22 |
| chr19_+_3572925 | 0.11 |
ENST00000333651.6
ENST00000417382.1 ENST00000453933.1 ENST00000262949.7 |
HMG20B
|
high mobility group 20B |
| chr19_+_38794797 | 0.11 |
ENST00000301246.5
ENST00000588605.1 |
C19orf33
|
chromosome 19 open reading frame 33 |
| chr1_+_9005917 | 0.11 |
ENST00000549778.1
ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6
|
carbonic anhydrase VI |
| chr6_-_88411911 | 0.10 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr11_-_66056478 | 0.10 |
ENST00000431556.2
ENST00000528575.1 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr17_-_39769005 | 0.10 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr19_-_54676884 | 0.10 |
ENST00000376591.4
|
TMC4
|
transmembrane channel-like 4 |
| chr22_+_31644388 | 0.10 |
ENST00000333611.4
ENST00000340552.4 |
LIMK2
|
LIM domain kinase 2 |
| chr22_+_31608219 | 0.09 |
ENST00000406516.1
ENST00000444929.2 ENST00000331728.4 |
LIMK2
|
LIM domain kinase 2 |
| chr19_-_46389359 | 0.08 |
ENST00000302165.3
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1 |
| chr3_+_32280159 | 0.08 |
ENST00000458535.2
ENST00000307526.3 |
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr17_+_40932610 | 0.08 |
ENST00000246914.5
|
WNK4
|
WNK lysine deficient protein kinase 4 |
| chr3_+_4535025 | 0.08 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr16_-_72698834 | 0.08 |
ENST00000570152.1
ENST00000561611.2 ENST00000570035.1 |
AC004158.2
|
AC004158.2 |
| chr11_-_6633799 | 0.07 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chrX_+_54466829 | 0.07 |
ENST00000375151.4
|
TSR2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr3_-_48632593 | 0.07 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr12_+_53773944 | 0.07 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr15_+_23810903 | 0.07 |
ENST00000564592.1
|
MKRN3
|
makorin ring finger protein 3 |
| chrX_+_49028265 | 0.07 |
ENST00000376322.3
ENST00000376327.5 |
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched) |
| chr2_+_241375069 | 0.07 |
ENST00000264039.2
|
GPC1
|
glypican 1 |
| chr7_-_129251459 | 0.07 |
ENST00000608694.1
|
RP11-448A19.1
|
RP11-448A19.1 |
| chr1_+_901847 | 0.07 |
ENST00000379410.3
ENST00000379409.2 ENST00000379407.3 |
PLEKHN1
|
pleckstrin homology domain containing, family N member 1 |
| chr12_-_68797580 | 0.07 |
ENST00000539404.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr2_+_38893047 | 0.06 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr2_-_177502254 | 0.06 |
ENST00000339037.3
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr11_+_63974135 | 0.06 |
ENST00000544997.1
ENST00000345728.5 ENST00000279227.5 |
FERMT3
|
fermitin family member 3 |
| chr2_+_74741569 | 0.06 |
ENST00000233638.7
|
TLX2
|
T-cell leukemia homeobox 2 |
| chr8_+_22250334 | 0.06 |
ENST00000520832.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr20_-_13619550 | 0.06 |
ENST00000455532.1
ENST00000544472.1 ENST00000539805.1 ENST00000337743.4 |
TASP1
|
taspase, threonine aspartase, 1 |
| chr19_-_10613862 | 0.06 |
ENST00000592055.1
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr19_+_49458107 | 0.06 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr1_-_32801825 | 0.06 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr11_-_118305921 | 0.06 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr11_-_61062762 | 0.06 |
ENST00000335613.5
|
VWCE
|
von Willebrand factor C and EGF domains |
| chr1_+_17248418 | 0.05 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr9_+_35605274 | 0.05 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chr22_+_38146987 | 0.05 |
ENST00000418339.1
|
TRIOBP
|
TRIO and F-actin binding protein |
| chr14_-_25519095 | 0.05 |
ENST00000419632.2
ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr12_-_75784669 | 0.05 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr3_+_51428704 | 0.05 |
ENST00000323686.4
|
RBM15B
|
RNA binding motif protein 15B |
| chr10_-_98273668 | 0.05 |
ENST00000357947.3
|
TLL2
|
tolloid-like 2 |
| chr20_-_25371412 | 0.05 |
ENST00000339157.5
ENST00000376542.3 |
ABHD12
|
abhydrolase domain containing 12 |
| chr11_-_65325430 | 0.05 |
ENST00000322147.4
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr19_+_36195429 | 0.05 |
ENST00000392197.2
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr17_+_3627185 | 0.04 |
ENST00000325418.4
|
GSG2
|
germ cell associated 2 (haspin) |
| chr1_+_43148625 | 0.04 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr10_-_50603497 | 0.04 |
ENST00000374139.2
|
DRGX
|
dorsal root ganglia homeobox |
| chr19_+_13229126 | 0.04 |
ENST00000292431.4
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chrX_+_51636629 | 0.04 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chrX_-_124097620 | 0.04 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr11_-_65624415 | 0.04 |
ENST00000524553.1
ENST00000527344.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr8_+_146277764 | 0.04 |
ENST00000331434.6
|
C8orf33
|
chromosome 8 open reading frame 33 |
| chr3_+_49591881 | 0.04 |
ENST00000296452.4
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr7_-_100065686 | 0.04 |
ENST00000423266.1
ENST00000456330.1 |
TSC22D4
|
TSC22 domain family, member 4 |
| chr19_+_45504688 | 0.04 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr1_-_11866034 | 0.03 |
ENST00000376590.3
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr17_+_48423453 | 0.03 |
ENST00000017003.2
ENST00000509778.1 ENST00000507602.1 |
XYLT2
|
xylosyltransferase II |
| chr14_-_25519317 | 0.03 |
ENST00000323944.5
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chrX_-_118826784 | 0.03 |
ENST00000394616.4
|
SEPT6
|
septin 6 |
| chr4_-_82393009 | 0.03 |
ENST00000436139.2
|
RASGEF1B
|
RasGEF domain family, member 1B |
| chr9_-_34662651 | 0.03 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr17_-_42188598 | 0.03 |
ENST00000591714.1
|
HDAC5
|
histone deacetylase 5 |
| chr19_+_4969116 | 0.03 |
ENST00000588337.1
ENST00000159111.4 ENST00000381759.4 |
KDM4B
|
lysine (K)-specific demethylase 4B |
| chr22_-_39096661 | 0.03 |
ENST00000216039.5
|
JOSD1
|
Josephin domain containing 1 |
| chr3_-_48130314 | 0.03 |
ENST00000439356.1
ENST00000395734.3 ENST00000426837.2 |
MAP4
|
microtubule-associated protein 4 |
| chr15_+_77287426 | 0.03 |
ENST00000558012.1
ENST00000267939.5 ENST00000379595.3 |
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr3_+_171757346 | 0.03 |
ENST00000421757.1
ENST00000415807.2 ENST00000392699.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr1_-_11865351 | 0.03 |
ENST00000413656.1
ENST00000376585.1 ENST00000423400.1 ENST00000431243.1 |
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr12_-_48213735 | 0.02 |
ENST00000417902.1
ENST00000417107.1 |
HDAC7
|
histone deacetylase 7 |
| chr12_-_48213568 | 0.02 |
ENST00000080059.7
ENST00000354334.3 ENST00000430670.1 ENST00000552960.1 ENST00000440293.1 |
HDAC7
|
histone deacetylase 7 |
| chr6_+_37787262 | 0.02 |
ENST00000287218.4
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr11_-_71752838 | 0.02 |
ENST00000537930.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr4_-_101439242 | 0.02 |
ENST00000296420.4
|
EMCN
|
endomucin |
| chr20_+_55205825 | 0.02 |
ENST00000544508.1
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr1_+_104068312 | 0.02 |
ENST00000524631.1
ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chrX_+_115567767 | 0.02 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr13_-_76056250 | 0.02 |
ENST00000377636.3
ENST00000431480.2 ENST00000377625.2 ENST00000425511.1 |
TBC1D4
|
TBC1 domain family, member 4 |
| chr12_+_53443963 | 0.02 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr2_-_37551846 | 0.02 |
ENST00000443187.1
|
PRKD3
|
protein kinase D3 |
| chr11_-_35440796 | 0.01 |
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr1_+_214163033 | 0.01 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr12_-_65153175 | 0.01 |
ENST00000543646.1
ENST00000542058.1 ENST00000258145.3 |
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr19_+_13228917 | 0.01 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr22_+_31644309 | 0.01 |
ENST00000425203.1
|
LIMK2
|
LIM domain kinase 2 |
| chr14_+_24837226 | 0.01 |
ENST00000554050.1
ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr10_-_98480243 | 0.01 |
ENST00000339364.5
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr13_-_27334879 | 0.01 |
ENST00000405846.3
|
GPR12
|
G protein-coupled receptor 12 |
| chr3_+_50192833 | 0.01 |
ENST00000426511.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr1_+_150122034 | 0.01 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr12_+_57984965 | 0.01 |
ENST00000540759.2
ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr4_+_154387480 | 0.01 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr10_+_102505954 | 0.01 |
ENST00000556085.1
ENST00000427256.1 |
PAX2
|
paired box 2 |
| chr6_+_42531798 | 0.00 |
ENST00000372903.2
ENST00000372899.1 ENST00000372901.1 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr2_+_232573208 | 0.00 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr11_-_71752571 | 0.00 |
ENST00000544238.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr11_+_66045634 | 0.00 |
ENST00000528852.1
ENST00000311445.6 |
CNIH2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr6_+_7108210 | 0.00 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr6_+_7107830 | 0.00 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr11_-_47869865 | 0.00 |
ENST00000530326.1
ENST00000532747.1 |
NUP160
|
nucleoporin 160kDa |
| chr2_+_232573222 | 0.00 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr11_+_64009072 | 0.00 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP2E
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0051511 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.0 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.0 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.1 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0032173 | septin ring(GO:0005940) septin collar(GO:0032173) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |