Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
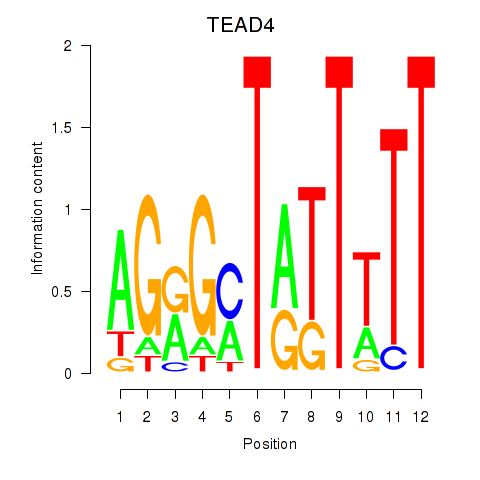
Results for TEAD4
Z-value: 0.86
Transcription factors associated with TEAD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD4
|
ENSG00000197905.4 | TEA domain transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD4 | hg19_v2_chr12_+_3069037_3069119 | -0.74 | 9.0e-02 | Click! |
Activity profile of TEAD4 motif
Sorted Z-values of TEAD4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_108905325 | 0.56 |
ENST00000438339.1
ENST00000409880.1 ENST00000437390.2 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr21_+_17909594 | 0.51 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr16_-_85969774 | 0.39 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr2_-_151395525 | 0.38 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr7_-_14029283 | 0.38 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr12_+_86268065 | 0.33 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chr3_+_184534994 | 0.31 |
ENST00000441141.1
ENST00000445089.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr13_+_113760098 | 0.30 |
ENST00000346342.3
ENST00000541084.1 ENST00000375581.3 |
F7
|
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr16_+_30386098 | 0.30 |
ENST00000322861.7
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr4_+_141264597 | 0.28 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr2_+_121493717 | 0.28 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr6_-_134638767 | 0.27 |
ENST00000524929.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr11_-_64570624 | 0.27 |
ENST00000439069.1
|
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr4_+_141178440 | 0.26 |
ENST00000394205.3
|
SCOC
|
short coiled-coil protein |
| chr5_-_108063949 | 0.26 |
ENST00000606054.1
|
LINC01023
|
long intergenic non-protein coding RNA 1023 |
| chr11_-_71639446 | 0.25 |
ENST00000534704.1
|
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr8_-_30002179 | 0.25 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr9_+_131902346 | 0.25 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chrX_+_49832231 | 0.25 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr7_-_111428957 | 0.24 |
ENST00000417165.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr3_-_12587055 | 0.24 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr14_+_76072096 | 0.23 |
ENST00000555058.1
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr2_+_196440692 | 0.23 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr6_-_41039567 | 0.23 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr2_+_145425534 | 0.22 |
ENST00000432608.1
ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr7_-_32529973 | 0.22 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr10_+_15074190 | 0.22 |
ENST00000428897.1
ENST00000413672.1 |
OLAH
|
oleoyl-ACP hydrolase |
| chr10_-_119805558 | 0.21 |
ENST00000369199.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr5_-_41794663 | 0.21 |
ENST00000510634.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr6_+_122720681 | 0.21 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr9_-_15472730 | 0.21 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr5_+_173472607 | 0.20 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr11_+_1295809 | 0.19 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr9_+_123906331 | 0.19 |
ENST00000431571.1
|
CNTRL
|
centriolin |
| chr1_+_174670143 | 0.18 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr4_+_78804393 | 0.18 |
ENST00000502384.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr6_+_96025341 | 0.18 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr9_+_42671887 | 0.17 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr9_+_67968793 | 0.17 |
ENST00000417488.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr2_+_239047337 | 0.17 |
ENST00000409223.1
ENST00000305959.4 |
KLHL30
|
kelch-like family member 30 |
| chrX_-_54824673 | 0.17 |
ENST00000218436.6
|
ITIH6
|
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
| chr1_+_241695670 | 0.17 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr3_-_69249863 | 0.17 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr21_+_34804479 | 0.17 |
ENST00000421802.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr4_-_47983519 | 0.16 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr6_+_131958436 | 0.16 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr4_-_110723335 | 0.16 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr3_-_196987309 | 0.16 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr4_-_39033963 | 0.15 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr2_+_152214098 | 0.15 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr17_-_38545799 | 0.15 |
ENST00000577541.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr12_+_20963632 | 0.15 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr8_-_70745575 | 0.15 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr15_-_83224682 | 0.14 |
ENST00000562833.1
|
RP11-152F13.10
|
RP11-152F13.10 |
| chr4_-_110723134 | 0.14 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr3_+_111805182 | 0.14 |
ENST00000430855.1
ENST00000431717.2 ENST00000264848.5 |
C3orf52
|
chromosome 3 open reading frame 52 |
| chr20_-_44516256 | 0.14 |
ENST00000372519.3
|
SPATA25
|
spermatogenesis associated 25 |
| chr1_+_241695424 | 0.14 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr11_-_59633951 | 0.14 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr12_+_20963647 | 0.14 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr14_+_76071805 | 0.13 |
ENST00000539311.1
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr7_-_82073109 | 0.13 |
ENST00000356860.3
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chrX_+_22056165 | 0.13 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr10_-_75410771 | 0.13 |
ENST00000372873.4
|
SYNPO2L
|
synaptopodin 2-like |
| chr22_-_46450024 | 0.13 |
ENST00000396008.2
ENST00000333761.1 |
C22orf26
|
chromosome 22 open reading frame 26 |
| chr2_-_133429091 | 0.13 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr12_+_69201923 | 0.12 |
ENST00000462284.1
ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr3_+_98699880 | 0.12 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr2_-_151344172 | 0.12 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr3_+_132316081 | 0.12 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr4_-_186317034 | 0.12 |
ENST00000505916.1
|
LRP2BP
|
LRP2 binding protein |
| chr6_-_109330702 | 0.12 |
ENST00000356644.7
|
SESN1
|
sestrin 1 |
| chr2_-_161056802 | 0.12 |
ENST00000283249.2
ENST00000409872.1 |
ITGB6
|
integrin, beta 6 |
| chr8_-_27115903 | 0.12 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr15_+_59908633 | 0.12 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr2_-_188419200 | 0.12 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr2_-_231989808 | 0.12 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr4_-_13546632 | 0.11 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr12_-_49333446 | 0.11 |
ENST00000537495.1
|
AC073610.5
|
Uncharacterized protein |
| chr8_+_24151553 | 0.11 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr9_-_96215822 | 0.11 |
ENST00000375412.5
|
FAM120AOS
|
family with sequence similarity 120A opposite strand |
| chr5_-_138862326 | 0.11 |
ENST00000330794.4
|
TMEM173
|
transmembrane protein 173 |
| chr11_-_104916034 | 0.11 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr3_-_142608001 | 0.11 |
ENST00000295992.3
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr1_+_171283331 | 0.11 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr19_+_37837218 | 0.11 |
ENST00000591134.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr4_-_110723194 | 0.10 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr3_+_111805262 | 0.10 |
ENST00000484828.1
|
C3orf52
|
chromosome 3 open reading frame 52 |
| chr3_+_42892458 | 0.10 |
ENST00000494619.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr16_+_53241854 | 0.10 |
ENST00000565803.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr11_+_120255997 | 0.10 |
ENST00000532993.1
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr19_-_3786354 | 0.10 |
ENST00000395040.2
ENST00000310132.6 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr1_+_90098606 | 0.10 |
ENST00000370454.4
|
LRRC8C
|
leucine rich repeat containing 8 family, member C |
| chr15_+_59730348 | 0.10 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr6_-_56507586 | 0.10 |
ENST00000439203.1
ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST
|
dystonin |
| chr7_+_120629653 | 0.10 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr21_+_17214724 | 0.10 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr8_+_55370487 | 0.10 |
ENST00000297316.4
|
SOX17
|
SRY (sex determining region Y)-box 17 |
| chr3_+_138067666 | 0.09 |
ENST00000475711.1
ENST00000464896.1 |
MRAS
|
muscle RAS oncogene homolog |
| chr8_-_134511587 | 0.09 |
ENST00000523855.1
ENST00000523854.1 |
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr13_+_78315528 | 0.09 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr8_+_107282368 | 0.09 |
ENST00000521369.2
|
RP11-395G23.3
|
RP11-395G23.3 |
| chrX_+_21392873 | 0.09 |
ENST00000379510.3
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr12_-_76478446 | 0.09 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr10_-_95462265 | 0.09 |
ENST00000536233.1
ENST00000359204.4 ENST00000371430.2 ENST00000394100.2 |
FRA10AC1
|
fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 |
| chr9_+_125273081 | 0.08 |
ENST00000335302.5
|
OR1J2
|
olfactory receptor, family 1, subfamily J, member 2 |
| chr19_+_35630628 | 0.08 |
ENST00000588715.1
ENST00000588607.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr1_+_107683436 | 0.08 |
ENST00000370068.1
|
NTNG1
|
netrin G1 |
| chrX_+_52240504 | 0.08 |
ENST00000399805.2
|
XAGE1B
|
X antigen family, member 1B |
| chr12_+_42725554 | 0.08 |
ENST00000546750.1
ENST00000547847.1 |
PPHLN1
|
periphilin 1 |
| chr9_-_33402506 | 0.08 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr14_-_89883412 | 0.08 |
ENST00000557258.1
|
FOXN3
|
forkhead box N3 |
| chr20_-_23402028 | 0.08 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr3_-_71179699 | 0.08 |
ENST00000497355.1
|
FOXP1
|
forkhead box P1 |
| chr3_-_52486841 | 0.08 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr12_-_123011476 | 0.08 |
ENST00000528279.1
ENST00000344591.4 ENST00000526560.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr18_-_3219847 | 0.08 |
ENST00000261606.7
|
MYOM1
|
myomesin 1 |
| chr8_+_92114060 | 0.07 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr10_-_735553 | 0.07 |
ENST00000280886.6
ENST00000423550.1 |
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr7_+_150811705 | 0.07 |
ENST00000335367.3
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr5_-_138775177 | 0.07 |
ENST00000302060.5
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr3_-_71179988 | 0.07 |
ENST00000491238.1
|
FOXP1
|
forkhead box P1 |
| chr2_-_188419078 | 0.07 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr15_+_100106155 | 0.07 |
ENST00000557785.1
ENST00000558049.1 ENST00000449277.2 |
MEF2A
|
myocyte enhancer factor 2A |
| chr9_-_34397800 | 0.07 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr10_+_5005445 | 0.07 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr4_-_89152474 | 0.07 |
ENST00000515655.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr13_-_46679144 | 0.07 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr17_+_57274914 | 0.07 |
ENST00000582004.1
ENST00000577660.1 |
PRR11
CTD-2510F5.6
|
proline rich 11 Uncharacterized protein |
| chr19_-_3786253 | 0.07 |
ENST00000585778.1
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr1_+_41204506 | 0.07 |
ENST00000525290.1
ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr6_-_152958521 | 0.06 |
ENST00000367255.5
ENST00000265368.4 ENST00000448038.1 ENST00000341594.5 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr4_-_100485183 | 0.06 |
ENST00000394876.2
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr2_-_161056762 | 0.06 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chr1_+_206623784 | 0.06 |
ENST00000426388.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr2_-_42588338 | 0.06 |
ENST00000234301.2
|
COX7A2L
|
cytochrome c oxidase subunit VIIa polypeptide 2 like |
| chr15_+_100106244 | 0.06 |
ENST00000557942.1
|
MEF2A
|
myocyte enhancer factor 2A |
| chr12_+_123011776 | 0.06 |
ENST00000450485.2
ENST00000333479.7 |
KNTC1
|
kinetochore associated 1 |
| chr13_+_20268547 | 0.06 |
ENST00000601204.1
|
AL354808.2
|
AL354808.2 |
| chr14_-_106791536 | 0.06 |
ENST00000390613.2
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr2_-_119605253 | 0.06 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr12_+_69864129 | 0.06 |
ENST00000547219.1
ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chr7_-_82073031 | 0.06 |
ENST00000356253.5
ENST00000423588.1 |
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr19_+_36346734 | 0.06 |
ENST00000586102.3
|
KIRREL2
|
kin of IRRE like 2 (Drosophila) |
| chr17_-_15175604 | 0.06 |
ENST00000431343.1
|
AC005703.3
|
AC005703.3 |
| chr5_+_138609782 | 0.06 |
ENST00000361059.2
ENST00000514694.1 ENST00000504203.1 ENST00000502929.1 ENST00000394800.2 ENST00000509644.1 ENST00000505016.1 |
MATR3
|
matrin 3 |
| chr10_-_4285923 | 0.06 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr2_+_38893047 | 0.06 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr9_+_125281420 | 0.06 |
ENST00000340750.1
|
OR1J4
|
olfactory receptor, family 1, subfamily J, member 4 |
| chr5_-_138861926 | 0.05 |
ENST00000510817.1
|
TMEM173
|
transmembrane protein 173 |
| chrX_-_52258669 | 0.05 |
ENST00000441417.1
|
XAGE1A
|
X antigen family, member 1A |
| chr16_-_46797149 | 0.05 |
ENST00000536476.1
|
MYLK3
|
myosin light chain kinase 3 |
| chr18_-_25616519 | 0.05 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr6_-_152489484 | 0.05 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr7_+_18535346 | 0.05 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chr11_-_92930556 | 0.05 |
ENST00000529184.1
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr21_+_17443521 | 0.05 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr2_-_20251744 | 0.05 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr9_+_2029019 | 0.05 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_38893208 | 0.05 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr1_-_175162048 | 0.05 |
ENST00000444639.1
|
KIAA0040
|
KIAA0040 |
| chr10_+_24528108 | 0.05 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr15_-_69113218 | 0.05 |
ENST00000560303.1
ENST00000465139.2 |
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chrX_-_15402498 | 0.05 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr17_+_32582293 | 0.05 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr3_+_189349162 | 0.05 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr17_+_29664830 | 0.04 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr11_-_71639613 | 0.04 |
ENST00000528184.1
ENST00000528511.2 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr6_-_134639180 | 0.04 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr18_+_3466248 | 0.04 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr7_-_22234381 | 0.04 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr1_+_46016703 | 0.04 |
ENST00000481885.1
ENST00000351829.4 ENST00000471651.1 |
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr5_-_41794313 | 0.04 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr6_-_32634425 | 0.04 |
ENST00000399082.3
ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr13_-_95131923 | 0.04 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr1_-_87379785 | 0.04 |
ENST00000401030.3
ENST00000370554.1 |
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr12_-_9913489 | 0.04 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr15_+_72947079 | 0.04 |
ENST00000421285.3
|
GOLGA6B
|
golgin A6 family, member B |
| chr4_+_120056939 | 0.04 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr7_+_123488124 | 0.04 |
ENST00000476325.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr21_-_44035168 | 0.04 |
ENST00000419628.1
|
AP001626.1
|
AP001626.1 |
| chr21_+_47878757 | 0.04 |
ENST00000400274.1
ENST00000427143.2 ENST00000318711.7 ENST00000457905.3 ENST00000466639.1 ENST00000435722.3 ENST00000417564.2 |
DIP2A
|
DIP2 disco-interacting protein 2 homolog A (Drosophila) |
| chr14_+_61201445 | 0.04 |
ENST00000261245.4
ENST00000539616.2 |
MNAT1
|
MNAT CDK-activating kinase assembly factor 1 |
| chr2_-_211341411 | 0.04 |
ENST00000233714.4
ENST00000443314.1 ENST00000441020.3 ENST00000450366.2 ENST00000431941.2 |
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr8_+_107282389 | 0.04 |
ENST00000577661.1
ENST00000445937.1 |
RP11-395G23.3
OXR1
|
RP11-395G23.3 oxidation resistance 1 |
| chr9_+_90497741 | 0.04 |
ENST00000325643.5
|
SPATA31E1
|
SPATA31 subfamily E, member 1 |
| chr2_-_179672142 | 0.04 |
ENST00000342992.6
ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN
|
titin |
| chr20_+_47538357 | 0.03 |
ENST00000371917.4
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr6_-_76782371 | 0.03 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr4_+_71588372 | 0.03 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr11_-_79151695 | 0.03 |
ENST00000278550.7
|
TENM4
|
teneurin transmembrane protein 4 |
| chr3_+_138327417 | 0.03 |
ENST00000338446.4
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr2_+_223916862 | 0.03 |
ENST00000604125.1
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4 |
| chr5_-_124081008 | 0.03 |
ENST00000306315.5
|
ZNF608
|
zinc finger protein 608 |
| chr6_-_13328564 | 0.03 |
ENST00000606530.1
ENST00000607658.1 ENST00000343141.4 ENST00000356436.4 ENST00000379300.3 ENST00000452989.1 ENST00000450347.1 ENST00000422136.1 ENST00000446018.1 ENST00000379291.1 ENST00000379307.2 ENST00000606370.1 ENST00000607230.1 |
TBC1D7
|
TBC1 domain family, member 7 |
| chr2_-_128568721 | 0.03 |
ENST00000322313.4
ENST00000393006.1 ENST00000409658.3 ENST00000436787.1 |
WDR33
|
WD repeat domain 33 |
| chr10_+_47894572 | 0.03 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr21_-_27423339 | 0.03 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr13_+_78315295 | 0.03 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chrX_-_10588595 | 0.03 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.5 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0042662 | endodermal cell fate determination(GO:0007493) negative regulation of mesodermal cell fate specification(GO:0042662) regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) gall bladder development(GO:0061010) |
| 0.0 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.0 | GO:0032279 | asymmetric synapse(GO:0032279) symmetric synapse(GO:0032280) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.1 | 0.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.2 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.2 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |