Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
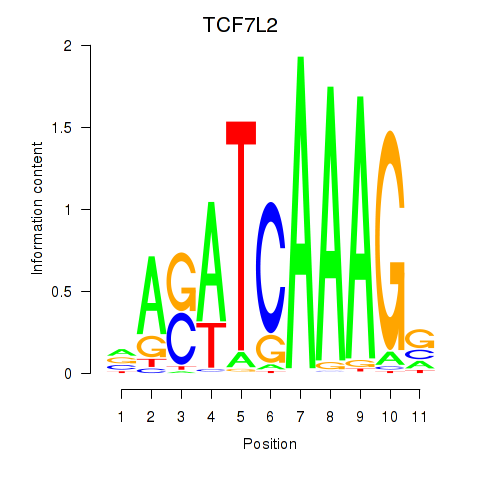
Results for TCF7L2
Z-value: 0.54
Transcription factors associated with TCF7L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L2
|
ENSG00000148737.11 | transcription factor 7 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7L2 | hg19_v2_chr10_+_114709999_114710031 | -0.80 | 5.4e-02 | Click! |
Activity profile of TCF7L2 motif
Sorted Z-values of TCF7L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_63557309 | 0.50 |
ENST00000580513.1
|
AXIN2
|
axin 2 |
| chr1_-_219615984 | 0.34 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr13_+_76334498 | 0.31 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr4_+_183065793 | 0.30 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr10_-_14613968 | 0.29 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr18_+_46065570 | 0.27 |
ENST00000591412.1
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr4_+_87857538 | 0.25 |
ENST00000511442.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr18_+_19668021 | 0.24 |
ENST00000579830.1
|
RP11-595B24.2
|
Uncharacterized protein |
| chr9_+_97766409 | 0.24 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr10_-_14614311 | 0.24 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr10_-_75351088 | 0.23 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr9_+_97766469 | 0.22 |
ENST00000433691.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr19_-_44008863 | 0.21 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr11_-_75921780 | 0.21 |
ENST00000529461.1
|
WNT11
|
wingless-type MMTV integration site family, member 11 |
| chr7_-_75452673 | 0.20 |
ENST00000416943.1
|
CCL24
|
chemokine (C-C motif) ligand 24 |
| chr11_-_76155618 | 0.20 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr5_+_133450365 | 0.18 |
ENST00000342854.5
ENST00000321603.6 ENST00000321584.4 ENST00000378564.1 ENST00000395029.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr1_-_11907829 | 0.17 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr10_-_104192405 | 0.17 |
ENST00000369937.4
|
CUEDC2
|
CUE domain containing 2 |
| chr3_-_134093395 | 0.17 |
ENST00000249883.5
|
AMOTL2
|
angiomotin like 2 |
| chr5_+_102200948 | 0.16 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr10_-_14614095 | 0.14 |
ENST00000482277.1
ENST00000378462.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr10_+_7745303 | 0.14 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr17_+_1674982 | 0.14 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr15_-_90294523 | 0.14 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chr5_+_170846640 | 0.13 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr16_+_53242350 | 0.13 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr3_-_134093275 | 0.13 |
ENST00000513145.1
ENST00000422605.2 |
AMOTL2
|
angiomotin like 2 |
| chr4_-_146859623 | 0.13 |
ENST00000379448.4
ENST00000513320.1 |
ZNF827
|
zinc finger protein 827 |
| chr10_+_7745232 | 0.13 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_-_14614122 | 0.13 |
ENST00000378465.3
ENST00000452706.2 ENST00000378458.2 |
FAM107B
|
family with sequence similarity 107, member B |
| chr14_+_62037287 | 0.13 |
ENST00000556569.1
|
RP11-47I22.3
|
Uncharacterized protein |
| chr11_+_45944190 | 0.13 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chrX_-_19765692 | 0.12 |
ENST00000432234.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr17_+_34848049 | 0.12 |
ENST00000588902.1
ENST00000591067.1 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr14_+_68189190 | 0.12 |
ENST00000539142.1
|
RDH12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis) |
| chr13_-_30160925 | 0.12 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr5_-_16742330 | 0.12 |
ENST00000505695.1
ENST00000427430.2 |
MYO10
|
myosin X |
| chr17_-_73389854 | 0.11 |
ENST00000578961.1
ENST00000392564.1 ENST00000582582.1 |
GRB2
|
growth factor receptor-bound protein 2 |
| chr22_-_33968239 | 0.11 |
ENST00000452586.2
ENST00000421768.1 |
LARGE
|
like-glycosyltransferase |
| chr14_-_23762777 | 0.11 |
ENST00000431326.2
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr3_-_134093738 | 0.11 |
ENST00000506107.1
|
AMOTL2
|
angiomotin like 2 |
| chr11_+_114128522 | 0.11 |
ENST00000535401.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr12_+_85673868 | 0.11 |
ENST00000316824.3
|
ALX1
|
ALX homeobox 1 |
| chr17_-_38256973 | 0.11 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr8_-_16859690 | 0.10 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr12_-_133464151 | 0.10 |
ENST00000315585.7
ENST00000266880.7 ENST00000443047.2 ENST00000432561.2 ENST00000450056.2 |
CHFR
|
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase |
| chr19_+_50084561 | 0.10 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr2_+_228678550 | 0.10 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr1_-_109935819 | 0.10 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr16_+_19222479 | 0.09 |
ENST00000568433.1
|
SYT17
|
synaptotagmin XVII |
| chr16_-_73082274 | 0.09 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr1_+_61330931 | 0.09 |
ENST00000371191.1
|
NFIA
|
nuclear factor I/A |
| chr22_+_19950060 | 0.09 |
ENST00000449653.1
|
COMT
|
catechol-O-methyltransferase |
| chr17_+_75447326 | 0.09 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr17_-_40169659 | 0.09 |
ENST00000457167.4
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr5_+_102201430 | 0.09 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr16_+_50582222 | 0.08 |
ENST00000268459.3
|
NKD1
|
naked cuticle homolog 1 (Drosophila) |
| chr11_+_65408273 | 0.08 |
ENST00000394227.3
|
SIPA1
|
signal-induced proliferation-associated 1 |
| chr6_-_28220002 | 0.08 |
ENST00000377294.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr1_-_40367668 | 0.08 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr13_+_76334795 | 0.08 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr7_-_148725733 | 0.08 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr4_-_69215699 | 0.08 |
ENST00000510746.1
ENST00000344157.4 ENST00000355665.3 |
YTHDC1
|
YTH domain containing 1 |
| chr17_-_63557759 | 0.08 |
ENST00000307078.5
|
AXIN2
|
axin 2 |
| chr13_+_76334567 | 0.08 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr7_+_73868439 | 0.07 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr14_+_75230011 | 0.07 |
ENST00000552421.1
ENST00000325680.7 ENST00000238571.3 |
YLPM1
|
YLP motif containing 1 |
| chr10_+_54074033 | 0.07 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr8_+_29953163 | 0.07 |
ENST00000518192.1
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr6_-_11779840 | 0.07 |
ENST00000506810.1
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr9_-_32635667 | 0.07 |
ENST00000242310.4
|
TAF1L
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 210kDa-like |
| chr12_+_6982725 | 0.07 |
ENST00000433346.1
|
LRRC23
|
leucine rich repeat containing 23 |
| chr2_+_69240415 | 0.07 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr2_+_69240511 | 0.07 |
ENST00000409349.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr11_+_7597639 | 0.07 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr5_-_135290705 | 0.07 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr17_-_39316983 | 0.07 |
ENST00000390661.3
|
KRTAP4-4
|
keratin associated protein 4-4 |
| chr4_-_157892055 | 0.06 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr6_-_32145861 | 0.06 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr5_-_16936340 | 0.06 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr2_+_47168630 | 0.06 |
ENST00000263737.6
|
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr7_-_27219849 | 0.06 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr10_-_105677427 | 0.06 |
ENST00000369764.1
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr17_-_46035187 | 0.06 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr9_-_15510287 | 0.06 |
ENST00000397519.2
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr2_+_177028805 | 0.06 |
ENST00000249440.3
|
HOXD3
|
homeobox D3 |
| chr5_+_60628074 | 0.06 |
ENST00000252744.5
|
ZSWIM6
|
zinc finger, SWIM-type containing 6 |
| chr6_-_135424186 | 0.06 |
ENST00000529882.1
|
HBS1L
|
HBS1-like (S. cerevisiae) |
| chr4_+_86525299 | 0.06 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr2_+_11679963 | 0.06 |
ENST00000263834.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr3_+_57875711 | 0.06 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr4_+_95972822 | 0.05 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr1_-_166028709 | 0.05 |
ENST00000595430.1
|
AL626787.1
|
AL626787.1 |
| chr4_+_170581213 | 0.05 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr5_-_160973649 | 0.05 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chrX_-_39956656 | 0.05 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chr5_+_175288631 | 0.05 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr1_-_115053781 | 0.05 |
ENST00000358465.2
ENST00000369543.2 |
TRIM33
|
tripartite motif containing 33 |
| chr1_+_151138526 | 0.05 |
ENST00000368902.1
|
SCNM1
|
sodium channel modifier 1 |
| chr2_+_171571827 | 0.05 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr11_-_62474803 | 0.05 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr12_+_54694979 | 0.05 |
ENST00000552848.1
|
COPZ1
|
coatomer protein complex, subunit zeta 1 |
| chr1_-_110933663 | 0.05 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr17_+_80818231 | 0.05 |
ENST00000576996.1
|
TBCD
|
tubulin folding cofactor D |
| chr8_+_55467072 | 0.05 |
ENST00000602362.1
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr2_-_9143786 | 0.05 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr3_+_57875738 | 0.05 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chrX_+_47004639 | 0.04 |
ENST00000345781.6
|
RBM10
|
RNA binding motif protein 10 |
| chr3_-_33686925 | 0.04 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr7_+_80275953 | 0.04 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr3_-_79068138 | 0.04 |
ENST00000495273.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr19_-_40791211 | 0.04 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr3_+_29323043 | 0.04 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr18_+_46065393 | 0.04 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr4_-_146859787 | 0.04 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr7_-_148581251 | 0.04 |
ENST00000478654.1
ENST00000460911.1 ENST00000350995.2 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr5_-_153857819 | 0.04 |
ENST00000231121.2
|
HAND1
|
heart and neural crest derivatives expressed 1 |
| chr7_+_27282319 | 0.04 |
ENST00000222761.3
|
EVX1
|
even-skipped homeobox 1 |
| chr2_+_173724771 | 0.04 |
ENST00000538974.1
ENST00000540783.1 |
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chrX_-_100183894 | 0.04 |
ENST00000328526.5
ENST00000372956.2 |
XKRX
|
XK, Kell blood group complex subunit-related, X-linked |
| chr6_-_24645956 | 0.04 |
ENST00000543707.1
|
KIAA0319
|
KIAA0319 |
| chr6_+_152130240 | 0.04 |
ENST00000427531.2
|
ESR1
|
estrogen receptor 1 |
| chr9_+_128510454 | 0.04 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr17_-_7297519 | 0.04 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chrX_+_47004599 | 0.04 |
ENST00000329236.7
|
RBM10
|
RNA binding motif protein 10 |
| chr1_+_162351503 | 0.03 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr6_-_11779403 | 0.03 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr10_-_101380121 | 0.03 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chrX_+_2670066 | 0.03 |
ENST00000381174.5
ENST00000419513.2 ENST00000426774.1 |
XG
|
Xg blood group |
| chr1_+_223101757 | 0.03 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr20_-_656437 | 0.03 |
ENST00000488788.2
|
RP5-850E9.3
|
Uncharacterized protein |
| chr17_+_79679299 | 0.03 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr2_-_80531399 | 0.03 |
ENST00000409148.1
ENST00000415098.1 ENST00000452811.1 |
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr17_+_48611853 | 0.03 |
ENST00000507709.1
ENST00000515126.1 ENST00000507467.1 |
EPN3
|
epsin 3 |
| chr14_-_81425828 | 0.03 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr7_-_16505440 | 0.03 |
ENST00000307068.4
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr15_-_65670360 | 0.03 |
ENST00000327987.4
|
IGDCC3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr6_-_112575758 | 0.03 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr4_-_109684120 | 0.03 |
ENST00000512646.1
ENST00000411864.2 ENST00000296486.3 ENST00000510706.1 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chrX_+_100646190 | 0.03 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr11_-_62572901 | 0.03 |
ENST00000439713.2
ENST00000531131.1 ENST00000530875.1 ENST00000531709.2 ENST00000294172.2 |
NXF1
|
nuclear RNA export factor 1 |
| chr17_+_42923686 | 0.03 |
ENST00000591513.1
|
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr1_+_62417957 | 0.03 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr17_-_56065540 | 0.03 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr7_+_80275752 | 0.03 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr7_-_105926058 | 0.03 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr8_-_142012169 | 0.03 |
ENST00000517453.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr14_-_21502944 | 0.03 |
ENST00000382951.3
|
RNASE13
|
ribonuclease, RNase A family, 13 (non-active) |
| chr17_-_73389737 | 0.03 |
ENST00000392563.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chr11_-_6677018 | 0.03 |
ENST00000299441.3
|
DCHS1
|
dachsous cadherin-related 1 |
| chrX_+_100645812 | 0.03 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr2_+_201936458 | 0.03 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr17_-_7297833 | 0.03 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr1_-_161993616 | 0.03 |
ENST00000294794.3
|
OLFML2B
|
olfactomedin-like 2B |
| chr1_+_147013182 | 0.03 |
ENST00000234739.3
|
BCL9
|
B-cell CLL/lymphoma 9 |
| chr9_+_139221880 | 0.03 |
ENST00000392945.3
ENST00000440944.1 |
GPSM1
|
G-protein signaling modulator 1 |
| chr6_+_35704855 | 0.02 |
ENST00000288065.2
ENST00000373866.3 |
ARMC12
|
armadillo repeat containing 12 |
| chr11_-_94965667 | 0.02 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr14_-_75179774 | 0.02 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr18_-_24445664 | 0.02 |
ENST00000578776.1
|
AQP4
|
aquaporin 4 |
| chr1_+_16693578 | 0.02 |
ENST00000401088.4
ENST00000471507.1 ENST00000401089.3 ENST00000375590.3 ENST00000492354.1 |
SZRD1
|
SUZ RNA binding domain containing 1 |
| chr1_-_94079648 | 0.02 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr17_+_37618257 | 0.02 |
ENST00000447079.4
|
CDK12
|
cyclin-dependent kinase 12 |
| chr15_+_90744745 | 0.02 |
ENST00000558051.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr6_-_11779174 | 0.02 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr14_+_32798462 | 0.02 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr7_-_148581360 | 0.02 |
ENST00000320356.2
ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr3_+_155838337 | 0.02 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr15_+_96897466 | 0.02 |
ENST00000558382.1
ENST00000558499.1 |
RP11-522B15.3
|
RP11-522B15.3 |
| chrX_+_106163626 | 0.02 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr17_-_56065484 | 0.02 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr12_-_31477072 | 0.02 |
ENST00000454658.2
|
FAM60A
|
family with sequence similarity 60, member A |
| chr6_+_64281906 | 0.02 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr5_+_102201509 | 0.02 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_+_154244987 | 0.02 |
ENST00000328703.7
ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr11_-_76155700 | 0.02 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr17_-_39123144 | 0.02 |
ENST00000355612.2
|
KRT39
|
keratin 39 |
| chr3_-_33686743 | 0.02 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr9_-_23826298 | 0.02 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr2_+_69240302 | 0.02 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr17_-_39093672 | 0.02 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr12_+_70760056 | 0.02 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr7_-_27213893 | 0.02 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr15_+_57511609 | 0.02 |
ENST00000543579.1
ENST00000537840.1 ENST00000343827.3 |
TCF12
|
transcription factor 12 |
| chr8_+_32579341 | 0.02 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr15_+_43985084 | 0.02 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr20_+_30555805 | 0.02 |
ENST00000562532.2
|
XKR7
|
XK, Kell blood group complex subunit-related family, member 7 |
| chr3_-_149375783 | 0.02 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr10_+_114710516 | 0.02 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr5_+_140235469 | 0.02 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr7_+_99613212 | 0.01 |
ENST00000426572.1
ENST00000535170.1 |
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr22_+_29279552 | 0.01 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr5_+_66300446 | 0.01 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr5_-_175815565 | 0.01 |
ENST00000509257.1
ENST00000507413.1 ENST00000510123.1 |
NOP16
|
NOP16 nucleolar protein |
| chr6_-_112575838 | 0.01 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr6_-_112575687 | 0.01 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr12_+_80603233 | 0.01 |
ENST00000547103.1
ENST00000458043.2 |
OTOGL
|
otogelin-like |
| chr3_+_141106643 | 0.01 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr6_+_37137939 | 0.01 |
ENST00000373509.5
|
PIM1
|
pim-1 oncogene |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7L2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.2 | GO:0100012 | negative regulation of mesodermal cell fate specification(GO:0042662) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.1 | 0.2 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 0.2 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.0 | 0.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.2 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.1 | GO:0045360 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.1 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0031337 | cellular response to phosphate starvation(GO:0016036) regulation of sulfur amino acid metabolic process(GO:0031335) positive regulation of sulfur amino acid metabolic process(GO:0031337) regulation of homocysteine metabolic process(GO:0050666) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.0 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.0 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.3 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.0 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |